List of verbs¶
Verbs are the building blocks of how you can use Miller to process your data. When you type
mlr --icsv --opprint sort -n quantity then head -n 4 example.csv
color shape flag k index quantity rate red circle true 3 16 13.8103 2.9010 yellow triangle true 1 11 43.6498 9.8870 yellow circle true 9 87 63.5058 8.3350 yellow circle true 8 73 63.9785 4.2370
the sort and head bits are verbs. See the Miller command
structure page for context.
At the command line, you can use mlr -l and mlr -L for information much
like what's on this page.
Overview¶
Whereas the Unix toolkit is made of the separate executables cat, tail, cut,
sort, etc., Miller has subcommands, or verbs, such as mlr cat, mlr tail, mlr cut, and
mlr sort, invoked as follows:
mlr tac *.dat mlr cut --complement -f os_version *.dat mlr sort -f hostname,uptime *.dat
These fall into categories as follows:
-
Analogs of their Unix-toolkit namesakes, discussed below as well as in Unix-toolkit Context: cat, cut, grep, head, join, sort, tac, tail, top, uniq.
-
awk-like functionality: filter, put, sec2gmt, sec2gmtdate, step, tee. -
Statistically oriented: bar, bootstrap, decimate, histogram, least-frequent, most-frequent, sample, shuffle, stats1, stats2.
-
Particularly oriented toward Record Heterogeneity, although all Miller commands can handle heterogeneous records: group-by, group-like, having-fields.
-
These draw from other sources (see also How Original Is Miller?): count-distinct is SQL-ish, and rename can be done by
sed(which does it faster: see Performance). Verbs: check, count-distinct, label, merge-fields, nest, nothing, regularize, rename, reorder, reshape, seqgen.
altkv¶
Map list of values to alternating key/value pairs.
mlr altkv -h
Usage: mlr altkv [options] Given fields with values of the form a,b,c,d,e,f emits a=b,c=d,e=f pairs. Options: -h|--help Show this message.
echo 'a,b,c,d,e,f' | mlr altkv
a=b,c=d,e=f
echo 'a,b,c,d,e,f,g' | mlr altkv
a=b,c=d,e=f,4=g
bar¶
Cheesy bar-charting.
mlr bar -h
Usage: mlr bar [options]
Replaces a numeric field with a number of asterisks, allowing for cheesy
bar plots. These align best with --opprint or --oxtab output format.
Options:
-f {a,b,c} Field names to convert to bars.
--lo {lo} Lower-limit value for min-width bar: default '0.000000'.
--hi {hi} Upper-limit value for max-width bar: default '100.000000'.
-w {n} Bar-field width: default '40'.
--auto Automatically computes limits, ignoring --lo and --hi.
Holds all records in memory before producing any output.
-c {character} Fill character: default '*'.
-x {character} Out-of-bounds character: default '#'.
-b {character} Blank character: default '.'.
Nominally the fill, out-of-bounds, and blank characters will be strings of length 1.
However you can make them all longer if you so desire.
-h|--help Show this message.
mlr --opprint cat data/small
a b i x y pan pan 1 0.346791 0.726802 eks pan 2 0.758679 0.522151 wye wye 3 0.204603 0.338318 eks wye 4 0.381399 0.134188 wye pan 5 0.573288 0.863624
mlr --opprint bar --lo 0 --hi 1 -f x,y data/small
a b i x y pan pan 1 *************........................... *****************************........... eks pan 2 ******************************.......... ********************.................... wye wye 3 ********................................ *************........................... eks wye 4 ***************......................... *****................................... wye pan 5 **********************.................. **********************************......
mlr --opprint bar --lo 0.4 --hi 0.6 -f x,y data/small
a b i x y pan pan 1 #....................................... ***************************************# eks pan 2 ***************************************# ************************................ wye wye 3 #....................................... #....................................... eks wye 4 #....................................... #....................................... wye pan 5 **********************************...... ***************************************#
mlr --opprint bar --auto -f x,y -w 20 data/small
a b i x y pan pan 1 [0.204603]*****...............[0.758679] [0.134188]****************....[0.863624] eks pan 2 [0.204603]*******************#[0.758679] [0.134188]**********..........[0.863624] wye wye 3 [0.204603]#...................[0.758679] [0.134188]*****...............[0.863624] eks wye 4 [0.204603]******..............[0.758679] [0.134188]#...................[0.863624] wye pan 5 [0.204603]*************.......[0.758679] [0.134188]*******************#[0.863624]
bootstrap¶
mlr bootstrap --help
Usage: mlr bootstrap [options]
Emits an n-sample, with replacement, of the input records.
See also mlr sample and mlr shuffle.
Options:
-n Number of samples to output. Defaults to number of input records.
Must be non-negative.
-h|--help Show this message.
The canonical use for bootstrap sampling is to put error bars on statistical quantities, such as mean. For example:
mlr --c2p stats1 -a mean,count -f u -g color data/colored-shapes.csv
color u_mean u_count yellow 0.4971291160651098 1413 red 0.49255964641241273 4641 purple 0.49400496322241666 1142 green 0.5048610595130744 1109 blue 0.5177171537414964 1470 orange 0.49053241584158375 303
mlr --c2p bootstrap then stats1 -a mean,count -f u -g color data/colored-shapes.csv
color u_mean u_count red 0.49183858109559747 4655 yellow 0.487271566995769 1418 green 0.5018994641860465 1075 orange 0.5005396620689654 290 blue 0.5309761257817928 1439 purple 0.4917481873438798 1201
color u_mean u_count
yellow 0.4809714157857651 1419 blue 0.5057790647530039 1498 red 0.49114305508382283 4593 purple 0.49652395202020194 1188 green 0.5011425433212993 1108 orange 0.48935696323529426 272
mlr --c2p bootstrap then stats1 -a mean,count -f u -g color data/colored-shapes.csv
color u_mean u_count red 0.49934473217726466 4671 purple 0.4934976176735793 1109 blue 0.5097866573146287 1497 yellow 0.4987188126740959 1436 orange 0.4802164827586204 290 green 0.5129018241860459 1075
case¶
mlr case --help
Usage: mlr case [options]
Uppercases strings in record keys and/or values.
Options:
-k Case only keys, not keys and values.
-v Case only values, not keys and values.
-f {a,b,c} Specify which field names to case (default: all)
-u Convert to uppercase
-l Convert to lowercase
-s Convert to sentence case (capitalize first letter)
-t Convert to title case (capitalize words)
-h|--help Show this message.
mlr --from test/input/cases.csv --icsv --ojson case -u
mlr --from test/input/cases.csv --icsv --ojson case -u -k
mlr --from test/input/cases.csv --icsv --ojson case -u -v
mlr --from test/input/cases.csv --icsv --ojson case -k -t then case -v -s
mlr --from test/input/cases.csv --icsv --ojson case -u -f apple,ball then case -l -f cat,dog
cat¶
Most useful for format conversions (see File Formats) and concatenating multiple same-schema CSV files to have the same header:
mlr cat -h
Usage: mlr cat [options]
Passes input records directly to output. Most useful for format conversion.
Options:
-n Prepend field "n" to each record with record-counter starting at 1.
-N {name} Prepend field {name} to each record with record-counter starting at 1.
-g {a,b,c} Optional group-by-field names for counters, e.g. a,b,c
--filename Prepend current filename to each record.
--filenum Prepend current filenum (1-up) to each record.
-h|--help Show this message.
cat data/a.csv
a,b,c 1,2,3 4,5,6
cat data/b.csv
a,b,c 7,8,9
mlr --csv cat data/a.csv data/b.csv
a,b,c 1,2,3 4,5,6 7,8,9
mlr --icsv --oxtab cat data/a.csv data/b.csv
a 1 b 2 c 3 a 4 b 5 c 6 a 7 b 8 c 9
mlr --csv cat -n data/a.csv data/b.csv
n,a,b,c 1,1,2,3 2,4,5,6 3,7,8,9
mlr --opprint cat data/small
a b i x y pan pan 1 0.346791 0.726802 eks pan 2 0.758679 0.522151 wye wye 3 0.204603 0.338318 eks wye 4 0.381399 0.134188 wye pan 5 0.573288 0.863624
mlr --opprint cat -n -g a data/small
n a b i x y 1 pan pan 1 0.346791 0.726802 1 eks pan 2 0.758679 0.522151 1 wye wye 3 0.204603 0.338318 2 eks wye 4 0.381399 0.134188 2 wye pan 5 0.573288 0.863624
check¶
mlr check --help
Usage: mlr check [options] Consumes records without printing any output, Useful for doing a well-formatted check on input data. with the exception that warnings are printed to stderr. Current checks are: * Data are parseable * If any key is the empty string Options: -h|--help Show this message.
clean-whitespace¶
mlr clean-whitespace --help
Usage: mlr clean-whitespace [options] For each record, for each field in the record, whitespace-cleans the keys and/or values. Whitespace-cleaning entails stripping leading and trailing whitespace, and replacing multiple whitespace with singles. For finer-grained control, please see the DSL functions lstrip, rstrip, strip, collapse_whitespace, and clean_whitespace. Options: -k|--keys-only Do not touch values. -v|--values-only Do not touch keys. It is an error to specify -k as well as -v -- to clean keys and values, leave off -k as well as -v. -h|--help Show this message.
mlr --icsv --ojson cat data/clean-whitespace.csv
[
{
" Name ": " Ann Simons",
" Preference ": " blue "
},
{
" Name ": "Bob Wang ",
" Preference ": " red "
},
{
" Name ": " Carol Vee",
" Preference ": " yellow"
}
]
mlr --icsv --ojson clean-whitespace -k data/clean-whitespace.csv
[
{
"Name": " Ann Simons",
"Preference": " blue "
},
{
"Name": "Bob Wang ",
"Preference": " red "
},
{
"Name": " Carol Vee",
"Preference": " yellow"
}
]
mlr --icsv --ojson clean-whitespace -v data/clean-whitespace.csv
[
{
" Name ": "Ann Simons",
" Preference ": "blue"
},
{
" Name ": "Bob Wang",
" Preference ": "red"
},
{
" Name ": "Carol Vee",
" Preference ": "yellow"
}
]
mlr --icsv --ojson clean-whitespace data/clean-whitespace.csv
[
{
"Name": "Ann Simons",
"Preference": "blue"
},
{
"Name": "Bob Wang",
"Preference": "red"
},
{
"Name": "Carol Vee",
"Preference": "yellow"
}
]
Function links:
count¶
mlr count --help
Usage: mlr count [options]
Prints number of records, optionally grouped by distinct values for specified field names.
Options:
-g {a,b,c} Optional group-by-field names for counts, e.g. a,b,c
-n {n} Show only the number of distinct values. Not interesting without -g.
-o {name} Field name for output-count. Default "count".
-h|--help Show this message.
mlr count data/medium
count=10000
mlr count -g a data/medium
a=pan,count=2081 a=eks,count=1965 a=wye,count=1966 a=zee,count=2047 a=hat,count=1941
mlr count -n -g a data/medium
count=5
mlr count -g b data/medium
b=pan,count=1942 b=wye,count=2057 b=zee,count=1943 b=eks,count=2008 b=hat,count=2050
mlr count -n -g b data/medium
count=5
mlr count -g a,b data/medium
a=pan,b=pan,count=427 a=eks,b=pan,count=371 a=wye,b=wye,count=377 a=eks,b=wye,count=407 a=wye,b=pan,count=392 a=zee,b=pan,count=389 a=eks,b=zee,count=357 a=zee,b=wye,count=455 a=hat,b=wye,count=423 a=pan,b=wye,count=395 a=zee,b=eks,count=391 a=hat,b=zee,count=385 a=hat,b=eks,count=389 a=wye,b=hat,count=426 a=pan,b=eks,count=429 a=eks,b=eks,count=413 a=hat,b=hat,count=381 a=hat,b=pan,count=363 a=zee,b=zee,count=403 a=pan,b=hat,count=417 a=pan,b=zee,count=413 a=zee,b=hat,count=409 a=wye,b=zee,count=385 a=eks,b=hat,count=417 a=wye,b=eks,count=386
count-distinct¶
mlr count-distinct --help
Usage: mlr count-distinct [options]
Prints number of records having distinct values for specified field names.
Same as uniq -c.
Options:
-f {a,b,c} Field names for distinct count.
-x {a,b,c} Field names to exclude for distinct count: use each record's others instead.
-n Show only the number of distinct values. Not compatible with -u.
-o {name} Field name for output count. Default "count".
Ignored with -u.
-u Do unlashed counts for multiple field names. With -f a,b and
without -u, computes counts for distinct combinations of a
and b field values. With -f a,b and with -u, computes counts
for distinct a field values and counts for distinct b field
values separately.
mlr count-distinct -f a,b then sort -nr count data/medium
a=zee,b=wye,count=455 a=pan,b=eks,count=429 a=pan,b=pan,count=427 a=wye,b=hat,count=426 a=hat,b=wye,count=423 a=pan,b=hat,count=417 a=eks,b=hat,count=417 a=eks,b=eks,count=413 a=pan,b=zee,count=413 a=zee,b=hat,count=409 a=eks,b=wye,count=407 a=zee,b=zee,count=403 a=pan,b=wye,count=395 a=wye,b=pan,count=392 a=zee,b=eks,count=391 a=zee,b=pan,count=389 a=hat,b=eks,count=389 a=wye,b=eks,count=386 a=hat,b=zee,count=385 a=wye,b=zee,count=385 a=hat,b=hat,count=381 a=wye,b=wye,count=377 a=eks,b=pan,count=371 a=hat,b=pan,count=363 a=eks,b=zee,count=357
mlr count-distinct -u -f a,b data/medium
field=a,value=pan,count=2081 field=a,value=eks,count=1965 field=a,value=wye,count=1966 field=a,value=zee,count=2047 field=a,value=hat,count=1941 field=b,value=pan,count=1942 field=b,value=wye,count=2057 field=b,value=zee,count=1943 field=b,value=eks,count=2008 field=b,value=hat,count=2050
mlr count-distinct -f a,b -o someothername then sort -nr someothername data/medium
a=zee,b=wye,someothername=455 a=pan,b=eks,someothername=429 a=pan,b=pan,someothername=427 a=wye,b=hat,someothername=426 a=hat,b=wye,someothername=423 a=pan,b=hat,someothername=417 a=eks,b=hat,someothername=417 a=eks,b=eks,someothername=413 a=pan,b=zee,someothername=413 a=zee,b=hat,someothername=409 a=eks,b=wye,someothername=407 a=zee,b=zee,someothername=403 a=pan,b=wye,someothername=395 a=wye,b=pan,someothername=392 a=zee,b=eks,someothername=391 a=zee,b=pan,someothername=389 a=hat,b=eks,someothername=389 a=wye,b=eks,someothername=386 a=hat,b=zee,someothername=385 a=wye,b=zee,someothername=385 a=hat,b=hat,someothername=381 a=wye,b=wye,someothername=377 a=eks,b=pan,someothername=371 a=hat,b=pan,someothername=363 a=eks,b=zee,someothername=357
mlr count-distinct -n -f a,b data/medium
count=25
count-similar¶
mlr count-similar --help
Usage: mlr count-similar [options]
Ingests all records, then emits each record augmented by a count of
the number of other records having the same group-by field values.
Options:
-g {a,b,c} Group-by-field names for counts, e.g. a,b,c
-o {name} Field name for output-counts. Defaults to "count".
-h|--help Show this message.
mlr --opprint head -n 20 data/medium
a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 zee pan 6 0.5271261600918548 0.49322128674835697 eks zee 7 0.6117840605678454 0.1878849191181694 zee wye 8 0.5985540091064224 0.976181385699006 hat wye 9 0.03144187646093577 0.7495507603507059 pan wye 10 0.5026260055412137 0.9526183602969864 pan pan 11 0.7930488423451967 0.6505816637259333 zee pan 12 0.3676141320555616 0.23614420670296965 eks pan 13 0.4915175580479536 0.7709126592971468 eks zee 14 0.5207382318405251 0.34141681118811673 eks pan 15 0.07155556372719507 0.3596137145616235 pan pan 16 0.5736853980681922 0.7554169353781729 zee eks 17 0.29081949506712723 0.054478717073354166 hat zee 18 0.05727869223575699 0.13343527626645157 zee pan 19 0.43144132839222604 0.8442204830496998 eks wye 20 0.38245149780530685 0.4730652428100751
mlr --opprint head -n 20 then count-similar -g a data/medium
a b i x y count pan pan 1 0.3467901443380824 0.7268028627434533 4 pan wye 10 0.5026260055412137 0.9526183602969864 4 pan pan 11 0.7930488423451967 0.6505816637259333 4 pan pan 16 0.5736853980681922 0.7554169353781729 4 eks pan 2 0.7586799647899636 0.5221511083334797 7 eks wye 4 0.38139939387114097 0.13418874328430463 7 eks zee 7 0.6117840605678454 0.1878849191181694 7 eks pan 13 0.4915175580479536 0.7709126592971468 7 eks zee 14 0.5207382318405251 0.34141681118811673 7 eks pan 15 0.07155556372719507 0.3596137145616235 7 eks wye 20 0.38245149780530685 0.4730652428100751 7 wye wye 3 0.20460330576630303 0.33831852551664776 2 wye pan 5 0.5732889198020006 0.8636244699032729 2 zee pan 6 0.5271261600918548 0.49322128674835697 5 zee wye 8 0.5985540091064224 0.976181385699006 5 zee pan 12 0.3676141320555616 0.23614420670296965 5 zee eks 17 0.29081949506712723 0.054478717073354166 5 zee pan 19 0.43144132839222604 0.8442204830496998 5 hat wye 9 0.03144187646093577 0.7495507603507059 2 hat zee 18 0.05727869223575699 0.13343527626645157 2
mlr --opprint head -n 20 then count-similar -g a then sort -f a data/medium
a b i x y count eks pan 2 0.7586799647899636 0.5221511083334797 7 eks wye 4 0.38139939387114097 0.13418874328430463 7 eks zee 7 0.6117840605678454 0.1878849191181694 7 eks pan 13 0.4915175580479536 0.7709126592971468 7 eks zee 14 0.5207382318405251 0.34141681118811673 7 eks pan 15 0.07155556372719507 0.3596137145616235 7 eks wye 20 0.38245149780530685 0.4730652428100751 7 hat wye 9 0.03144187646093577 0.7495507603507059 2 hat zee 18 0.05727869223575699 0.13343527626645157 2 pan pan 1 0.3467901443380824 0.7268028627434533 4 pan wye 10 0.5026260055412137 0.9526183602969864 4 pan pan 11 0.7930488423451967 0.6505816637259333 4 pan pan 16 0.5736853980681922 0.7554169353781729 4 wye wye 3 0.20460330576630303 0.33831852551664776 2 wye pan 5 0.5732889198020006 0.8636244699032729 2 zee pan 6 0.5271261600918548 0.49322128674835697 5 zee wye 8 0.5985540091064224 0.976181385699006 5 zee pan 12 0.3676141320555616 0.23614420670296965 5 zee eks 17 0.29081949506712723 0.054478717073354166 5 zee pan 19 0.43144132839222604 0.8442204830496998 5
cut¶
mlr cut --help
Usage: mlr cut [options]
Passes through input records with specified fields included/excluded.
Options:
-f {a,b,c} Comma-separated field names for cut, e.g. a,b,c.
-o Retain fields in the order specified here in the argument list.
Default is to retain them in the order found in the input data.
-x|--complement Exclude, rather than include, field names specified by -f.
-r Treat field names as regular expressions. "ab", "a.*b" will
match any field name containing the substring "ab" or matching
"a.*b", respectively; anchors of the form "^ab$", "^a.*b$" may
be used. The -o flag is ignored when -r is present.
-h|--help Show this message.
Examples:
mlr cut -f hostname,status
mlr cut -x -f hostname,status
mlr cut -r -f '^status$,sda[0-9]'
mlr cut -r -f '^status$,"sda[0-9]"'
mlr cut -r -f '^status$,"sda[0-9]"i' (this is case-insensitive)
mlr --opprint cat data/small
a b i x y pan pan 1 0.346791 0.726802 eks pan 2 0.758679 0.522151 wye wye 3 0.204603 0.338318 eks wye 4 0.381399 0.134188 wye pan 5 0.573288 0.863624
mlr --opprint cut -f y,x,i data/small
i x y 1 0.346791 0.726802 2 0.758679 0.522151 3 0.204603 0.338318 4 0.381399 0.134188 5 0.573288 0.863624
echo 'a=1,b=2,c=3' | mlr cut -f b,c,a
a=1,b=2,c=3
echo 'a=1,b=2,c=3' | mlr cut -o -f b,c,a
b=2,c=3,a=1
decimate¶
mlr decimate --help
Usage: mlr decimate [options]
Passes through one of every n records, optionally by category.
Options:
-b Decimate by printing first of every n.
-e Decimate by printing last of every n (default).
-g {a,b,c} Optional group-by-field names for decimate counts, e.g. a,b,c.
-n {n} Decimation factor (default 10).
-h|--help Show this message.
fill-down¶
mlr fill-down --help
Usage: mlr fill-down [options]
If a given record has a missing value for a given field, fill that from
the corresponding value from a previous record, if any.
By default, a 'missing' field either is absent, or has the empty-string value.
With -a, a field is 'missing' only if it is absent.
Options:
--all Operate on all fields in the input.
-a|--only-if-absent If a given record has a missing value for a given field,
fill that from the corresponding value from a previous record, if any.
By default, a 'missing' field either is absent, or has the empty-string value.
With -a, a field is 'missing' only if it is absent.
-f Field names for fill-down.
-h|--help Show this message.
cat data/fillable.csv
a,b,c 1,,3 4,5,6 7,,9
mlr --csv fill-down -f b data/fillable.csv
a,b,c 1,,3 4,5,6 7,5,9
mlr --csv fill-down -a -f b data/fillable.csv
a,b,c 1,,3 4,5,6 7,,9
fill-empty¶
mlr fill-empty --help
Usage: mlr fill-empty [options]
Fills empty-string fields with specified fill-value.
Options:
-v {string} Fill-value: defaults to "N/A"
-S Don't infer type -- so '-v 0' would fill string 0 not int 0.
cat data/fillable.csv
a,b,c 1,,3 4,5,6 7,,9
mlr --csv fill-empty data/fillable.csv
a,b,c 1,N/A,3 4,5,6 7,N/A,9
mlr --csv fill-empty -v something data/fillable.csv
a,b,c 1,something,3 4,5,6 7,something,9
filter¶
mlr filter --help
Usage: mlr filter [options] {DSL expression}
Lets you use a domain-specific language to progamatically filter which
stream records will be output.
See also: https://miller.readthedocs.io/en/latest/reference-verbs
Options:
-f {file name} File containing a DSL expression (see examples below). If the filename
is a directory, all *.mlr files in that directory are loaded.
-e {expression} You can use this after -f to add an expression. Example use
case: define functions/subroutines in a file you specify with -f, then call
them with an expression you specify with -e.
(If you mix -e and -f then the expressions are evaluated in the order encountered.
Since the expression pieces are simply concatenated, please be sure to use intervening
semicolons to separate expressions.)
-s name=value: Predefines out-of-stream variable @name to have
Thus mlr put -s foo=97 '$column += @foo' is like
mlr put 'begin {@foo = 97} $column += @foo'.
The value part is subject to type-inferencing.
May be specified more than once, e.g. -s name1=value1 -s name2=value2.
Note: the value may be an environment variable, e.g. -s sequence=$SEQUENCE
-x (default false) Prints records for which {expression} evaluates to false, not true,
i.e. invert the sense of the filter expression.
-q Does not include the modified record in the output stream.
Useful for when all desired output is in begin and/or end blocks.
-S and -F: There are no-ops in Miller 6 and above, since now type-inferencing is done
by the record-readers before filter/put is executed. Supported as no-op pass-through
flags for backward compatibility.
-h|--help Show this message.
Parser-info options:
-w Print warnings about things like uninitialized variables.
-W Same as -w, but exit the process if there are any warnings.
-p Prints the expressions's AST (abstract syntax tree), which gives full
transparency on the precedence and associativity rules of Miller's grammar,
to stdout.
-d Like -p but uses a parenthesized-expression format for the AST.
-D Like -d but with output all on one line.
-E Echo DSL expression before printing parse-tree
-v Same as -E -p.
-X Exit after parsing but before stream-processing. Useful with -v/-d/-D, if you
only want to look at parser information.
Records will pass the filter depending on the last bare-boolean statement in
the DSL expression. That can be the result of <, ==, >, etc., the return value of a function call
which returns boolean, etc.
Examples:
mlr --csv --from example.csv filter '$color == "red"'
mlr --csv --from example.csv filter '$color == "red" && flag == true'
More example filter expressions:
First record in each file:
'FNR == 1'
Subsampling:
'urand() < 0.001'
Compound booleans:
'$color != "blue" && $value > 4.2'
'($x < 0.5 && $y < 0.5) || ($x > 0.5 && $y > 0.5)'
Regexes with case-insensitive flag
'($name =~ "^sys.*east$") || ($name =~ "^dev.[0-9]+"i)'
Assignments, then bare-boolean filter statement:
'$ab = $a+$b; $cd = $c+$d; $ab != $cd'
Bare-boolean filter statement within a conditional:
'if (NR < 100) {
$x > 0.3;
} else {
$x > 0.002;
}
'
Using 'any' higher-order function to see if $index is 10, 20, or 30:
'any([10,20,30], func(e) {return $index == e})'
See also https://miller.readthedocs.io/reference-dsl for more context.
Features which filter shares with put¶
Please see DSL reference for more information about the expression language for mlr filter.
flatten¶
mlr flatten --help
Usage: mlr flatten [options]
Flattens multi-level maps to single-level ones. Example: field with name 'a'
and value '{"b": { "c": 4 }}' becomes name 'a.b.c' and value 4.
Options:
-f Comma-separated list of field names to flatten (default all).
-s Separator, defaulting to mlr --flatsep value.
-h|--help Show this message.
format-values¶
mlr format-values --help
Usage: mlr format-values [options]
Applies format strings to all field values, depending on autodetected type.
* If a field value is detected to be integer, applies integer format.
* Else, if a field value is detected to be float, applies float format.
* Else, applies string format.
Note: this is a low-keystroke way to apply formatting to many fields. To get
finer control, please see the fmtnum function within the mlr put DSL.
Note: this verb lets you apply arbitrary format strings, which can produce
undefined behavior and/or program crashes. See your system's "man printf".
Options:
-i {integer format} Defaults to "%d".
Examples: "%06lld", "%08llx".
Note that Miller integers are long long so you must use
formats which apply to long long, e.g. with ll in them.
Undefined behavior results otherwise.
-f {float format} Defaults to "%f".
Examples: "%8.3lf", "%.6le".
Note that Miller floats are double-precision so you must
use formats which apply to double, e.g. with l[efg] in them.
Undefined behavior results otherwise.
-s {string format} Defaults to "%s".
Examples: "_%s", "%08s".
Note that you must use formats which apply to string, e.g.
with s in them. Undefined behavior results otherwise.
-n Coerce field values autodetected as int to float, and then
apply the float format.
mlr --opprint format-values data/small
a b i x y pan pan 1 0.346791 0.726802 eks pan 2 0.758679 0.522151 wye wye 3 0.204603 0.338318 eks wye 4 0.381399 0.134188 wye pan 5 0.573288 0.863624
mlr --opprint format-values -n data/small
a b i x y pan pan 1.000000 0.346791 0.726802 eks pan 2.000000 0.758679 0.522151 wye wye 3.000000 0.204603 0.338318 eks wye 4.000000 0.381399 0.134188 wye pan 5.000000 0.573288 0.863624
mlr --opprint format-values -i %08llx -f %.6le -s X%sX data/small
a b i x y XpanX XpanX 00000001 3.467910e-01 7.268020e-01 XeksX XpanX 00000002 7.586790e-01 5.221510e-01 XwyeX XwyeX 00000003 2.046030e-01 3.383180e-01 XeksX XwyeX 00000004 3.813990e-01 1.341880e-01 XwyeX XpanX 00000005 5.732880e-01 8.636240e-01
mlr --opprint format-values -i %08llx -f %.6le -s X%sX -n data/small
a b i x y XpanX XpanX 1.000000e+00 3.467910e-01 7.268020e-01 XeksX XpanX 2.000000e+00 7.586790e-01 5.221510e-01 XwyeX XwyeX 3.000000e+00 2.046030e-01 3.383180e-01 XeksX XwyeX 4.000000e+00 3.813990e-01 1.341880e-01 XwyeX XpanX 5.000000e+00 5.732880e-01 8.636240e-01
fraction¶
mlr fraction --help
Usage: mlr fraction [options]
For each record's value in specified fields, computes the ratio of that
value to the sum of values in that field over all input records.
E.g. with input records x=1 x=2 x=3 and x=4, emits output records
x=1,x_fraction=0.1 x=2,x_fraction=0.2 x=3,x_fraction=0.3 and x=4,x_fraction=0.4
Note: this is internally a two-pass algorithm: on the first pass it retains
input records and accumulates sums; on the second pass it computes quotients
and emits output records. This means it produces no output until all input is read.
Options:
-f {a,b,c} Field name(s) for fraction calculation
-g {d,e,f} Optional group-by-field name(s) for fraction counts
-p Produce percents [0..100], not fractions [0..1]. Output field names
end with "_percent" rather than "_fraction"
-c Produce cumulative distributions, i.e. running sums: each output
value folds in the sum of the previous for the specified group
E.g. with input records x=1 x=2 x=3 and x=4, emits output records
x=1,x_cumulative_fraction=0.1 x=2,x_cumulative_fraction=0.3
x=3,x_cumulative_fraction=0.6 and x=4,x_cumulative_fraction=1.0
For example, suppose you have the following CSV file:
u=female,v=red,n=2458 u=female,v=green,n=192 u=female,v=blue,n=337 u=female,v=purple,n=468 u=female,v=yellow,n=3 u=female,v=orange,n=17 u=male,v=red,n=143 u=male,v=green,n=227 u=male,v=blue,n=2034 u=male,v=purple,n=12 u=male,v=yellow,n=1192 u=male,v=orange,n=448
Then we can see what each record's n contributes to the total n:
mlr --opprint fraction -f n data/fraction-example.csv
u v n n_fraction female red 2458 0.32638427831629263 female green 192 0.025494622228123754 female blue 337 0.04474837338998805 female purple 468 0.06214314168105165 female yellow 3 0.00039835347231443366 female orange 17 0.002257336343115124 male red 143 0.018988182180321337 male green 227 0.03014207940512548 male blue 2034 0.270083654229186 male purple 12 0.0015934138892577346 male yellow 1192 0.15827911299960165 male orange 448 0.0594874518656221
Using -g we can split those out by gender, or by color:
mlr --opprint fraction -f n -g u data/fraction-example.csv
u v n n_fraction female red 2458 0.7073381294964028 female green 192 0.05525179856115108 female blue 337 0.09697841726618706 female purple 468 0.13467625899280575 female yellow 3 0.0008633093525179857 female orange 17 0.004892086330935252 male red 143 0.035256410256410256 male green 227 0.05596646942800789 male blue 2034 0.5014792899408284 male purple 12 0.0029585798816568047 male yellow 1192 0.2938856015779093 male orange 448 0.11045364891518737
mlr --opprint fraction -f n -g v data/fraction-example.csv
u v n n_fraction female red 2458 0.9450211457131872 female green 192 0.45823389021479716 female blue 337 0.1421341206242092 female purple 468 0.975 female yellow 3 0.002510460251046025 female orange 17 0.03655913978494624 male red 143 0.05497885428681276 male green 227 0.5417661097852029 male blue 2034 0.8578658793757908 male purple 12 0.025 male yellow 1192 0.9974895397489539 male orange 448 0.9634408602150538
We can see, for example, that 70.9% of females have red (on the left) while 94.5% of reds are for females.
To convert fractions to percents, you may use -p:
mlr --opprint fraction -f n -p data/fraction-example.csv
u v n n_percent female red 2458 32.638427831629265 female green 192 2.5494622228123753 female blue 337 4.474837338998805 female purple 468 6.214314168105165 female yellow 3 0.039835347231443365 female orange 17 0.2257336343115124 male red 143 1.8988182180321338 male green 227 3.014207940512548 male blue 2034 27.0083654229186 male purple 12 0.15934138892577346 male yellow 1192 15.827911299960165 male orange 448 5.94874518656221
Another often-used idiom is to convert from a point distribution to a cumulative distribution, also known as "running sums". Here, you can use -c:
mlr --opprint fraction -f n -p -c data/fraction-example.csv
u v n n_cumulative_percent female red 2458 32.638427831629265 female green 192 35.18789005444164 female blue 337 39.66272739344044 female purple 468 45.87704156154561 female yellow 3 45.916876908777056 female orange 17 46.142610543088566 male red 143 48.041428761120706 male green 227 51.05563670163325 male blue 2034 78.06400212455186 male purple 12 78.22334351347763 male yellow 1192 94.0512548134378 male orange 448 100
mlr --opprint fraction -f n -g u -p -c data/fraction-example.csv
u v n n_cumulative_percent female red 2458 70.73381294964028 female green 192 76.2589928057554 female blue 337 85.9568345323741 female purple 468 99.42446043165467 female yellow 3 99.51079136690647 female orange 17 100 male red 143 3.5256410256410255 male green 227 9.122287968441814 male blue 2034 59.27021696252466 male purple 12 59.56607495069034 male yellow 1192 88.95463510848126 male orange 448 100
gap¶
mlr gap -h
Usage: mlr gap [options]
Emits an empty record every n records, or when certain values change.
Options:
Emits an empty record every n records, or when certain values change.
-g {a,b,c} Print a gap whenever values of these fields (e.g. a,b,c) changes.
-n {n} Print a gap every n records.
One of -f or -g is required.
-n is ignored if -g is present.
-h|--help Show this message.
grep¶
mlr grep -h
Usage: mlr grep [options] {regular expression}
Passes through records which match the regular expression.
Options:
-i Use case-insensitive search.
-v Invert: pass through records which do not match the regex.
-a Only grep for values, not keys and values.
-h|--help Show this message.
Note that "mlr filter" is more powerful, but requires you to know field names.
By contrast, "mlr grep" allows you to regex-match the entire record. It does this
by formatting each record in memory as DKVP (or NIDX, if -a is supplied), using
OFS "," and OPS "=", and matching the resulting line against the regex specified
here. In particular, the regex is not applied to the input stream: if you have
CSV with header line "x,y,z" and data line "1,2,3" then the regex will be
matched, not against either of these lines, but against the DKVP line
"x=1,y=2,z=3". Furthermore, not all the options to system grep are supported,
and this command is intended to be merely a keystroke-saver. To get all the
features of system grep, you can do
"mlr --odkvp ... | grep ... | mlr --idkvp ..."
group-by¶
mlr group-by --help
Usage: mlr group-by [options] {comma-separated field names}
Outputs records in batches having identical values at specified field names.Options:
-h|--help Show this message.
This is similar to sort but with less work. Namely, Miller's sort has three steps: read through the data and append linked lists of records, one for each unique combination of the key-field values; after all records are read, sort the key-field values; then print each record-list. The group-by operation simply omits the middle sort. An example should make this more clear:
mlr --opprint sort -f a data/small
a b i x y eks pan 2 0.758679 0.522151 eks wye 4 0.381399 0.134188 pan pan 1 0.346791 0.726802 wye wye 3 0.204603 0.338318 wye pan 5 0.573288 0.863624
mlr --opprint group-by a data/small
a b i x y pan pan 1 0.346791 0.726802 eks pan 2 0.758679 0.522151 eks wye 4 0.381399 0.134188 wye wye 3 0.204603 0.338318 wye pan 5 0.573288 0.863624
In this example, since the sort is on field a, the first step is to group together all records having the same value for field a; the second step is to sort the distinct a-field values pan, eks, and wye into eks, pan, and wye; the third step is to print out the record-list for a=eks, then the record-list for a=pan, then the record-list for a=wye. The group-by operation omits the middle sort and just puts like records together, for those times when a sort isn't desired. In particular, the ordering of group-by fields for group-by is the order in which they were encountered in the data stream, which in some cases may be more interesting to you.
group-like¶
mlr group-like --help
Usage: mlr group-like [options] Outputs records in batches having identical field names. Options: -h|--help Show this message.
This groups together records having the same schema (i.e. same ordered list of field names) which is useful for making sense of time-ordered output as described in Record Heterogeneity -- in particular, in preparation for CSV or pretty-print output.
mlr cat data/het.dkvp
resource=/path/to/file,loadsec=0.45,ok=true record_count=100,resource=/path/to/file resource=/path/to/second/file,loadsec=0.32,ok=true record_count=150,resource=/path/to/second/file resource=/some/other/path,loadsec=0.97,ok=false
mlr --opprint group-like data/het.dkvp
resource loadsec ok /path/to/file 0.45 true /path/to/second/file 0.32 true /some/other/path 0.97 false record_count resource 100 /path/to/file 150 /path/to/second/file
gsub¶
mlr gsub -h
Usage: mlr gsub [options]
Replaces old string with new string in specified field(s), with regex support
for the old string and handling multiple matches, like the `gsub` DSL function.
See also the `sub` and `ssub` verbs.
Options:
-f {a,b,c} Field names to convert.
-h|--help Show this message.
mlr --icsv --opprint --from example.csv cat --filename then sub -f color,shape l X
filename color shape flag k index quantity rate example.csv yeXlow triangXe true 1 11 43.6498 9.8870 example.csv red square true 2 15 79.2778 0.0130 example.csv red circXe true 3 16 13.8103 2.9010 example.csv red square false 4 48 77.5542 7.4670 example.csv purpXe triangXe false 5 51 81.2290 8.5910 example.csv red square false 6 64 77.1991 9.5310 example.csv purpXe triangXe false 7 65 80.1405 5.8240 example.csv yeXlow circXe true 8 73 63.9785 4.2370 example.csv yeXlow circXe true 9 87 63.5058 8.3350 example.csv purpXe square false 10 91 72.3735 8.2430
mlr --icsv --opprint --from example.csv cat --filename then gsub -f color,shape l X
filename color shape flag k index quantity rate example.csv yeXXow triangXe true 1 11 43.6498 9.8870 example.csv red square true 2 15 79.2778 0.0130 example.csv red circXe true 3 16 13.8103 2.9010 example.csv red square false 4 48 77.5542 7.4670 example.csv purpXe triangXe false 5 51 81.2290 8.5910 example.csv red square false 6 64 77.1991 9.5310 example.csv purpXe triangXe false 7 65 80.1405 5.8240 example.csv yeXXow circXe true 8 73 63.9785 4.2370 example.csv yeXXow circXe true 9 87 63.5058 8.3350 example.csv purpXe square false 10 91 72.3735 8.2430
having-fields¶
mlr having-fields --help
Usage: mlr having-fields [options]
Conditionally passes through records depending on each record's field names.
Options:
--at-least {comma-separated names}
--which-are {comma-separated names}
--at-most {comma-separated names}
--all-matching {regular expression}
--any-matching {regular expression}
--none-matching {regular expression}
Examples:
mlr having-fields --which-are amount,status,owner
mlr having-fields --any-matching 'sda[0-9]'
mlr having-fields --any-matching '"sda[0-9]"'
mlr having-fields --any-matching '"sda[0-9]"i' (this is case-insensitive)
Similar to group-like, this retains records with specified schema.
mlr cat data/het.dkvp
resource=/path/to/file,loadsec=0.45,ok=true record_count=100,resource=/path/to/file resource=/path/to/second/file,loadsec=0.32,ok=true record_count=150,resource=/path/to/second/file resource=/some/other/path,loadsec=0.97,ok=false
mlr having-fields --at-least resource data/het.dkvp
resource=/path/to/file,loadsec=0.45,ok=true record_count=100,resource=/path/to/file resource=/path/to/second/file,loadsec=0.32,ok=true record_count=150,resource=/path/to/second/file resource=/some/other/path,loadsec=0.97,ok=false
mlr having-fields --which-are resource,ok,loadsec data/het.dkvp
resource=/path/to/file,loadsec=0.45,ok=true resource=/path/to/second/file,loadsec=0.32,ok=true resource=/some/other/path,loadsec=0.97,ok=false
head¶
mlr head --help
Usage: mlr head [options]
Passes through the first n records, optionally by category.
Without -g, ceases consuming more input (i.e. is fast) when n records have been read.
Options:
-g {a,b,c} Optional group-by-field names for head counts, e.g. a,b,c.
-n {n} Head-count to print. Default 10.
-h|--help Show this message.
Note that head is distinct from top -- head shows fields which appear first in the data stream; top shows fields which are numerically largest (or smallest).
mlr --opprint head -n 4 data/medium
a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463
mlr --opprint head -n 1 -g b data/medium
a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 wye wye 3 0.20460330576630303 0.33831852551664776 eks zee 7 0.6117840605678454 0.1878849191181694 zee eks 17 0.29081949506712723 0.054478717073354166 wye hat 24 0.7286126830627567 0.19441962592638418
histogram¶
mlr histogram --help
Just a histogram. Input values < lo or > hi are not counted.
Usage: mlr histogram [options]
-f {a,b,c} Value-field names for histogram counts
--lo {lo} Histogram low value
--hi {hi} Histogram high value
--nbins {n} Number of histogram bins. Defaults to 20.
--auto Automatically computes limits, ignoring --lo and --hi.
Holds all values in memory before producing any output.
-o {prefix} Prefix for output field name. Default: no prefix.
-h|--help Show this message.
This is just a histogram; there's not too much to say here. A note about binning, by example: Suppose you use --lo 0.0 --hi 1.0 --nbins 10 -f x. The input numbers less than 0 or greater than 1 aren't counted in any bin. Input numbers equal to 1 are counted in the last bin. That is, bin 0 has 0.0 < x < 0.1, bin 1 has 0.1 < x < 0.2, etc., but bin 9 has 0.9 < x < 1.0.
mlr --opprint put '$x2=$x**2;$x3=$x2*$x' \ then histogram -f x,x2,x3 --lo 0 --hi 1 --nbins 10 \ data/medium
bin_lo bin_hi x_count x2_count x3_count 0 0.1 1072 3231 4661 0.1 0.2 938 1254 1184 0.2 0.3 1037 988 845 0.3 0.4 988 832 676 0.4 0.5 950 774 576 0.5 0.6 1002 692 476 0.6 0.7 1007 591 438 0.7 0.8 1007 560 420 0.8 0.9 986 571 383 0.9 1 1013 507 341
mlr --opprint put '$x2=$x**2;$x3=$x2*$x' \ then histogram -f x,x2,x3 --lo 0 --hi 1 --nbins 10 -o my_ \ data/medium
my_bin_lo my_bin_hi my_x_count my_x2_count my_x3_count 0 0.1 1072 3231 4661 0.1 0.2 938 1254 1184 0.2 0.3 1037 988 845 0.3 0.4 988 832 676 0.4 0.5 950 774 576 0.5 0.6 1002 692 476 0.6 0.7 1007 591 438 0.7 0.8 1007 560 420 0.8 0.9 986 571 383 0.9 1 1013 507 341
join¶
mlr join --help
Usage: mlr join [options]
Joins records from specified left file name with records from all file names
at the end of the Miller argument list.
Functionality is essentially the same as the system "join" command, but for
record streams.
Options:
-f {left file name}
-j {a,b,c} Comma-separated join-field names for output
-l {a,b,c} Comma-separated join-field names for left input file;
defaults to -j values if omitted.
-r {a,b,c} Comma-separated join-field names for right input file(s);
defaults to -j values if omitted.
--lk|--left-keep-field-names {a,b,c} If supplied, this means keep only the specified field
names from the left file. Automatically includes the join-field name(s). Helpful
for when you only want a limited subset of information from the left file.
Tip: you can use --lk "": this means the left file becomes solely a row-selector
for the input files.
--lp {text} Additional prefix for non-join output field names from
the left file
--rp {text} Additional prefix for non-join output field names from
the right file(s)
--np Do not emit paired records
--ul Emit unpaired records from the left file
--ur Emit unpaired records from the right file(s)
-s|--sorted-input Require sorted input: records must be sorted
lexically by their join-field names, else not all records will
be paired. The only likely use case for this is with a left
file which is too big to fit into system memory otherwise.
-u Enable unsorted input. (This is the default even without -u.)
In this case, the entire left file will be loaded into memory.
--prepipe {command} As in main input options; see mlr --help for details.
If you wish to use a prepipe command for the main input as well
as here, it must be specified there as well as here.
--prepipex {command} Likewise.
File-format options default to those for the right file names on the Miller
argument list, but may be overridden for the left file as follows. Please see
the main "mlr --help" for more information on syntax for these arguments:
-i {one of csv,dkvp,nidx,pprint,xtab}
--irs {record-separator character}
--ifs {field-separator character}
--ips {pair-separator character}
--repifs
--implicit-csv-header
--implicit-tsv-header
--no-implicit-csv-header
--no-implicit-tsv-header
For example, if you have 'mlr --csv ... join -l foo ... ' then the left-file format will
be specified CSV as well unless you override with 'mlr --csv ... join --ijson -l foo' etc.
Likewise, if you have 'mlr --csv --implicit-csv-header ...' then the join-in file will be
expected to be headerless as well unless you put '--no-implicit-csv-header' after 'join'.
Please use "mlr --usage-separator-options" for information on specifying separators.
Please see https://miller.readthedocs.io/en/latest/reference-verbs.html#join for more information
including examples.
Examples:
Join larger table with IDs with smaller ID-to-name lookup table, showing only paired records:
mlr --icsvlite --opprint cat data/join-left-example.csv
id name 100 alice 200 bob 300 carol 400 david 500 edgar
mlr --icsvlite --opprint cat data/join-right-example.csv
status idcode present 400 present 100 missing 200 present 100 present 200 missing 100 missing 200 present 300 missing 600 present 400 present 400 present 300 present 100 missing 400 present 200 present 200 present 200 present 200 present 400 present 300
mlr --icsvlite --opprint \ join -u -j id -r idcode -f data/join-left-example.csv \ data/join-right-example.csv
id name status 400 david present 100 alice present 200 bob missing 100 alice present 200 bob present 100 alice missing 200 bob missing 300 carol present 400 david present 400 david present 300 carol present 100 alice present 400 david missing 200 bob present 200 bob present 200 bob present 200 bob present 400 david present 300 carol present
Same, but with sorting the input first:
mlr --icsvlite --opprint sort -f idcode \ then join -j id -r idcode -f data/join-left-example.csv \ data/join-right-example.csv
id name status 100 alice present 100 alice present 100 alice missing 100 alice present 200 bob missing 200 bob present 200 bob missing 200 bob present 200 bob present 200 bob present 200 bob present 300 carol present 300 carol present 300 carol present 400 david present 400 david present 400 david present 400 david missing 400 david present
Same, but showing only unpaired records:
mlr --icsvlite --opprint \ join --np --ul --ur -u -j id -r idcode -f data/join-left-example.csv \ data/join-right-example.csv
status idcode missing 600 id name 500 edgar
Use prefixing options to disambiguate between otherwise identical non-join field names:
mlr --csvlite --opprint cat data/self-join.csv data/self-join.csv
a b c 1 2 3 1 4 5 1 2 3 1 4 5
mlr --csvlite --opprint join -j a --lp left_ --rp right_ -f data/self-join.csv data/self-join.csv
a left_b left_c right_b right_c 1 2 3 2 3 1 4 5 2 3 1 2 3 4 5 1 4 5 4 5
Use zero join columns:
mlr --csvlite --opprint join -j "" --lp left_ --rp right_ -f data/self-join.csv data/self-join.csv
left_a left_b left_c right_a right_b right_c 1 2 3 1 2 3 1 4 5 1 2 3 1 2 3 1 4 5 1 4 5 1 4 5
json-parse¶
mlr json-parse --help
Usage: mlr json-parse [options]
Tries to convert string field values to parsed JSON, e.g. "[1,2,3]" -> [1,2,3].
Options:
-f {...} Comma-separated list of field names to json-parse (default all).
-k If supplied, then on parse fail for any cell, keep the (unparsable)
input value for the cell.
-h|--help Show this message.
json-stringify¶
mlr json-stringify --help
Usage: mlr json-stringify [options]
Produces string field values from field-value data, e.g. [1,2,3] -> "[1,2,3]".
Options:
-f {...} Comma-separated list of field names to json-parse (default all).
--jvstack Produce multi-line JSON output.
--no-jvstack Produce single-line JSON output per record (default).
-h|--help Show this message.
label¶
mlr label --help
Usage: mlr label [options] {new1,new2,new3,...}
Given n comma-separated names, renames the first n fields of each record to
have the respective name. (Fields past the nth are left with their original
names.) Particularly useful with --inidx or --implicit-csv-header, to give
useful names to otherwise integer-indexed fields.
Options:
-h|--help Show this message.
See also rename.
Example: Files such as /etc/passwd, /etc/group, and so on have implicit field names which are found in section-5 manpages. These field names may be made explicit as follows:
% grep -v '^#' /etc/passwd | mlr --nidx --fs : --opprint label name,password,uid,gid,gecos,home_dir,shell | head name password uid gid gecos home_dir shell nobody * -2 -2 Unprivileged User /var/empty /usr/bin/false root * 0 0 System Administrator /var/root /bin/sh daemon * 1 1 System Services /var/root /usr/bin/false _uucp * 4 4 Unix to Unix Copy Protocol /var/spool/uucp /usr/sbin/uucico _taskgated * 13 13 Task Gate Daemon /var/empty /usr/bin/false _networkd * 24 24 Network Services /var/networkd /usr/bin/false _installassistant * 25 25 Install Assistant /var/empty /usr/bin/false _lp * 26 26 Printing Services /var/spool/cups /usr/bin/false _postfix * 27 27 Postfix Mail Server /var/spool/postfix /usr/bin/false
Likewise, if you have CSV/CSV-lite input data which has somehow been bereft of its header line, you can re-add a header line using --implicit-csv-header and label:
cat data/headerless.csv
John,23,present Fred,34,present Alice,56,missing Carol,45,present
mlr --csv --implicit-csv-header cat data/headerless.csv
1,2,3 John,23,present Fred,34,present Alice,56,missing Carol,45,present
mlr --csv --implicit-csv-header label name,age,status data/headerless.csv
name,age,status John,23,present Fred,34,present Alice,56,missing Carol,45,present
mlr --icsv --implicit-csv-header --opprint label name,age,status data/headerless.csv
name age status John 23 present Fred 34 present Alice 56 missing Carol 45 present
latin1-to-utf8¶
mlr latin1-to-utf8 -h
Usage: mlr latin1-to-utf8, with no options. Recursively converts record strings from Latin-1 to UTF-8. For field-level control, please see the latin1_to_utf8 DSL function. Options: -h|--help Show this message.
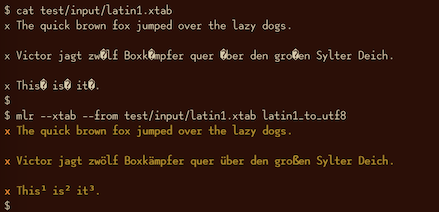
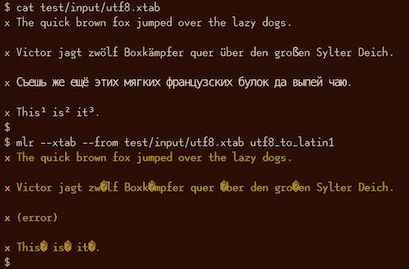
utf8-to-latin1¶
mlr utf8-to-latin1 -h
Usage: mlr utf8-to-latin1, with no options. Recursively converts record strings from Latin-1 to UTF-8. For field-level control, please see the utf8_to_latin1 DSL function. Options: -h|--help Show this message.
In this example, the English and German pangrams are convertible from UTF-8 to Latin-1, but the Russian one is not:
least-frequent¶
mlr least-frequent -h
Usage: mlr least-frequent [options]
Shows the least frequently occurring distinct values for specified field names.
The first entry is the statistical anti-mode; the remaining are runners-up.
Options:
-f {one or more comma-separated field names}. Required flag.
-n {count}. Optional flag defaulting to 10.
-b Suppress counts; show only field values.
-o {name} Field name for output count. Default "count".
See also "mlr most-frequent".
mlr --c2p --from data/colored-shapes.csv least-frequent -f shape -n 5
shape count circle 2591 triangle 3372 square 4115
mlr --c2p --from data/colored-shapes.csv least-frequent -f shape,color -n 5
shape color count circle orange 68 triangle orange 107 square orange 128 circle green 287 circle purple 289
mlr --c2p --from data/colored-shapes.csv least-frequent -f shape,color -n 5 -o someothername
shape color someothername circle orange 68 triangle orange 107 square orange 128 circle green 287 circle purple 289
mlr --c2p --from data/colored-shapes.csv least-frequent -f shape,color -n 5 -b
shape color circle orange triangle orange square orange circle green circle purple
See also most-frequent.
merge-fields¶
mlr merge-fields --help
Usage: mlr merge-fields [options]
Computes univariate statistics for each input record, accumulated across
specified fields.
Options:
-a {sum,count,...} Names of accumulators. One or more of:
count Count instances of fields
null_count Count number of empty-string/JSON-null instances per field
distinct_count Count number of distinct values per field
mode Find most-frequently-occurring values for fields; first-found wins tie
antimode Find least-frequently-occurring values for fields; first-found wins tie
sum Compute sums of specified fields
mean Compute averages (sample means) of specified fields
var Compute sample variance of specified fields
stddev Compute sample standard deviation of specified fields
meaneb Estimate error bars for averages (assuming no sample autocorrelation)
skewness Compute sample skewness of specified fields
kurtosis Compute sample kurtosis of specified fields
min Compute minimum values of specified fields
max Compute maximum values of specified fields
minlen Compute minimum string-lengths of specified fields
maxlen Compute maximum string-lengths of specified fields
-f {a,b,c} Value-field names on which to compute statistics. Requires -o.
-r {a,b,c} Regular expressions for value-field names on which to compute
statistics. Requires -o.
-c {a,b,c} Substrings for collapse mode. All fields which have the same names
after removing substrings will be accumulated together. Please see
examples below.
-i Use interpolated percentiles, like R's type=7; default like type=1.
Not sensical for string-valued fields.
-o {name} Output field basename for -f/-r.
-k Keep the input fields which contributed to the output statistics;
the default is to omit them.
String-valued data make sense unless arithmetic on them is required,
e.g. for sum, mean, interpolated percentiles, etc. In case of mixed data,
numbers are less than strings.
Example input data: "a_in_x=1,a_out_x=2,b_in_y=4,b_out_x=8".
Example: mlr merge-fields -a sum,count -f a_in_x,a_out_x -o foo
produces "b_in_y=4,b_out_x=8,foo_sum=3,foo_count=2" since "a_in_x,a_out_x" are
summed over.
Example: mlr merge-fields -a sum,count -r in_,out_ -o bar
produces "bar_sum=15,bar_count=4" since all four fields are summed over.
Example: mlr merge-fields -a sum,count -c in_,out_
produces "a_x_sum=3,a_x_count=2,b_y_sum=4,b_y_count=1,b_x_sum=8,b_x_count=1"
since "a_in_x" and "a_out_x" both collapse to "a_x", "b_in_y" collapses to
"b_y", and "b_out_x" collapses to "b_x".
This is like mlr stats1 but all accumulation is done across fields within each given record: horizontal rather than vertical statistics, if you will.
Examples:
mlr --csvlite --opprint cat data/inout.csv
a_in a_out b_in b_out 436 490 446 195 526 320 963 780 220 888 705 831
mlr --csvlite --opprint merge-fields -a min,max,sum -c _in,_out data/inout.csv
a_min a_max a_sum b_min b_max b_sum 436 490 926 195 446 641 320 526 846 780 963 1743 220 888 1108 705 831 1536
mlr --csvlite --opprint merge-fields -k -a sum -c _in,_out data/inout.csv
a_in a_out b_in b_out a_sum b_sum 436 490 446 195 926 641 526 320 963 780 846 1743 220 888 705 831 1108 1536
most-frequent¶
mlr most-frequent -h
Usage: mlr most-frequent [options]
Shows the most frequently occurring distinct values for specified field names.
The first entry is the statistical mode; the remaining are runners-up.
Options:
-f {one or more comma-separated field names}. Required flag.
-n {count}. Optional flag defaulting to 10.
-b Suppress counts; show only field values.
-o {name} Field name for output count. Default "count".
See also "mlr least-frequent".
mlr --c2p --from data/colored-shapes.csv most-frequent -f shape -n 5
shape count square 4115 triangle 3372 circle 2591
mlr --c2p --from data/colored-shapes.csv most-frequent -f shape,color -n 5
shape color count square red 1874 triangle red 1560 circle red 1207 square blue 589 square yellow 589
mlr --c2p --from data/colored-shapes.csv most-frequent -f shape,color -n 5 -o someothername
shape color someothername square red 1874 triangle red 1560 circle red 1207 square blue 589 square yellow 589
mlr --c2p --from data/colored-shapes.csv most-frequent -f shape,color -n 5 -b
shape color square red triangle red circle red square blue square yellow
See also least-frequent.
nest¶
mlr nest -h
Usage: mlr nest [options]
Explodes specified field values into separate fields/records, or reverses this.
Options:
--explode,--implode One is required.
--values,--pairs One is required.
--across-records,--across-fields One is required.
-f {field name} Required.
--nested-fs {string} Defaults to ";". Field separator for nested values.
--nested-ps {string} Defaults to ":". Pair separator for nested key-value pairs.
--evar {string} Shorthand for --explode --values --across-records --nested-fs {string}
--ivar {string} Shorthand for --implode --values --across-records --nested-fs {string}
Please use "mlr --usage-separator-options" for information on specifying separators.
Examples:
mlr nest --explode --values --across-records -f x
with input record "x=a;b;c,y=d" produces output records
"x=a,y=d"
"x=b,y=d"
"x=c,y=d"
Use --implode to do the reverse.
mlr nest --explode --values --across-fields -f x
with input record "x=a;b;c,y=d" produces output records
"x_1=a,x_2=b,x_3=c,y=d"
Use --implode to do the reverse.
mlr nest --explode --pairs --across-records -f x
with input record "x=a:1;b:2;c:3,y=d" produces output records
"a=1,y=d"
"b=2,y=d"
"c=3,y=d"
mlr nest --explode --pairs --across-fields -f x
with input record "x=a:1;b:2;c:3,y=d" produces output records
"a=1,b=2,c=3,y=d"
Notes:
* With --pairs, --implode doesn't make sense since the original field name has
been lost.
* The combination "--implode --values --across-records" is non-streaming:
no output records are produced until all input records have been read. In
particular, this means it won't work in `tail -f` contexts. But all other flag
combinations result in streaming (`tail -f` friendly) data processing.
If input is coming from `tail -f`, be sure to use `--records-per-batch 1`.
* It's up to you to ensure that the nested-fs is distinct from your data's IFS:
e.g. by default the former is semicolon and the latter is comma.
See also mlr reshape.
nothing¶
mlr nothing -h
Usage: mlr nothing [options] Drops all input records. Useful for testing, or after tee/print/etc. have produced other output. Options: -h|--help Show this message.
put¶
mlr put --help
Usage: mlr put [options] {DSL expression}
Lets you use a domain-specific language to progamatically alter stream records.
See also: https://miller.readthedocs.io/en/latest/reference-verbs
Options:
-f {file name} File containing a DSL expression (see examples below). If the filename
is a directory, all *.mlr files in that directory are loaded.
-e {expression} You can use this after -f to add an expression. Example use
case: define functions/subroutines in a file you specify with -f, then call
them with an expression you specify with -e.
(If you mix -e and -f then the expressions are evaluated in the order encountered.
Since the expression pieces are simply concatenated, please be sure to use intervening
semicolons to separate expressions.)
-s name=value: Predefines out-of-stream variable @name to have
Thus mlr put -s foo=97 '$column += @foo' is like
mlr put 'begin {@foo = 97} $column += @foo'.
The value part is subject to type-inferencing.
May be specified more than once, e.g. -s name1=value1 -s name2=value2.
Note: the value may be an environment variable, e.g. -s sequence=$SEQUENCE
-x (default false) Prints records for which {expression} evaluates to false, not true,
i.e. invert the sense of the filter expression.
-q Does not include the modified record in the output stream.
Useful for when all desired output is in begin and/or end blocks.
-S and -F: There are no-ops in Miller 6 and above, since now type-inferencing is done
by the record-readers before filter/put is executed. Supported as no-op pass-through
flags for backward compatibility.
-h|--help Show this message.
Parser-info options:
-w Print warnings about things like uninitialized variables.
-W Same as -w, but exit the process if there are any warnings.
-p Prints the expressions's AST (abstract syntax tree), which gives full
transparency on the precedence and associativity rules of Miller's grammar,
to stdout.
-d Like -p but uses a parenthesized-expression format for the AST.
-D Like -d but with output all on one line.
-E Echo DSL expression before printing parse-tree
-v Same as -E -p.
-X Exit after parsing but before stream-processing. Useful with -v/-d/-D, if you
only want to look at parser information.
Examples:
mlr --from example.csv put '$qr = $quantity * $rate'
More example put expressions:
If-statements:
'if ($flag == true) { $quantity *= 10}'
'if ($x > 0.0) { $y=log10($x); $z=sqrt($y) } else {$y = 0.0; $z = 0.0}'
Newly created fields can be read after being written:
'$new_field = $index**2; $qn = $quantity * $new_field'
Regex-replacement:
'$name = sub($name, "http.*com"i, "")'
Regex-capture:
'if ($a =~ "([a-z]+)_([0-9]+)") { $b = "left_\1"; $c = "right_\2" }'
Built-in variables:
'$filename = FILENAME'
Aggregations (use mlr put -q):
'@sum += $x; end {emit @sum}'
'@sum[$shape] += $quantity; end {emit @sum, "shape"}'
'@sum[$shape][$color] += $x; end {emit @sum, "shape", "color"}'
'
@min = min(@min,$x);
@max=max(@max,$x);
end{emitf @min, @max}
'
See also https://miller.readthedocs.io/reference-dsl for more context.
Features which put shares with filter¶
Please see the DSL reference for more information about the expression language for mlr put.
regularize¶
mlr regularize --help
Usage: mlr regularize [options] Outputs records sorted lexically ascending by keys. Options: -h|--help Show this message.
This exists since hash-map software in various languages and tools encountered in the wild does not always print similar rows with fields in the same order: mlr regularize helps clean that up.
See also reorder.
remove-empty-columns¶
mlr remove-empty-columns --help
Usage: mlr remove-empty-columns [options] Omits fields which are empty on every input row. Non-streaming. Options: -h|--help Show this message.
cat data/remove-empty-columns.csv
a,b,c,d,e 1,,3,,5 2,,4,,5 3,,5,,7
mlr --csv remove-empty-columns data/remove-empty-columns.csv
a,c,e 1,3,5 2,4,5 3,5,7
Since this verb needs to read all records to see if any of them has a non-empty value for a given field name, it is non-streaming: it will ingest all records before writing any.
rename¶
mlr rename --help
Usage: mlr rename [options] {old1,new1,old2,new2,...}
Renames specified fields.
Options:
-r Treat old field names as regular expressions. "ab", "a.*b"
will match any field name containing the substring "ab" or
matching "a.*b", respectively; anchors of the form "^ab$",
"^a.*b$" may be used. New field names may be plain strings,
or may contain capture groups of the form "\1" through
"\9". Wrapping the regex in double quotes is optional, but
is required if you wish to follow it with 'i' to indicate
case-insensitivity.
-g Do global replacement within each field name rather than
first-match replacement.
-h|--help Show this message.
Examples:
mlr rename old_name,new_name'
mlr rename old_name_1,new_name_1,old_name_2,new_name_2'
mlr rename -r 'Date_[0-9]+,Date,' Rename all such fields to be "Date"
mlr rename -r '"Date_[0-9]+",Date' Same
mlr rename -r 'Date_([0-9]+).*,\1' Rename all such fields to be of the form 20151015
mlr rename -r '"name"i,Name' Rename "name", "Name", "NAME", etc. to "Name"
mlr --opprint cat data/small
a b i x y pan pan 1 0.346791 0.726802 eks pan 2 0.758679 0.522151 wye wye 3 0.204603 0.338318 eks wye 4 0.381399 0.134188 wye pan 5 0.573288 0.863624
mlr --opprint rename i,INDEX,b,COLUMN2 data/small
a COLUMN2 INDEX x y pan pan 1 0.346791 0.726802 eks pan 2 0.758679 0.522151 wye wye 3 0.204603 0.338318 eks wye 4 0.381399 0.134188 wye pan 5 0.573288 0.863624
As discussed in Performance, sed is significantly faster than Miller at doing this. However, Miller is format-aware, so it knows to do renames only within specified field keys and not any others, nor in field values which may happen to contain the same pattern. Example:
sed 's/y/COLUMN5/g' data/small
a=pan,b=pan,i=1,x=0.346791,COLUMN5=0.726802 a=eks,b=pan,i=2,x=0.758679,COLUMN5=0.522151 a=wCOLUMN5e,b=wCOLUMN5e,i=3,x=0.204603,COLUMN5=0.338318 a=eks,b=wCOLUMN5e,i=4,x=0.381399,COLUMN5=0.134188 a=wCOLUMN5e,b=pan,i=5,x=0.573288,COLUMN5=0.863624
mlr rename y,COLUMN5 data/small
a=pan,b=pan,i=1,x=0.346791,COLUMN5=0.726802 a=eks,b=pan,i=2,x=0.758679,COLUMN5=0.522151 a=wye,b=wye,i=3,x=0.204603,COLUMN5=0.338318 a=eks,b=wye,i=4,x=0.381399,COLUMN5=0.134188 a=wye,b=pan,i=5,x=0.573288,COLUMN5=0.863624
See also label.
reorder¶
mlr reorder --help
Usage: mlr reorder [options]
Moves specified names to start of record, or end of record.
Options:
-e Put specified field names at record end: default is to put them at record start.
-f {a,b,c} Field names to reorder.
-b {x} Put field names specified with -f before field name specified by {x},
if any. If {x} isn't present in a given record, the specified fields
will not be moved.
-a {x} Put field names specified with -f after field name specified by {x},
if any. If {x} isn't present in a given record, the specified fields
will not be moved.
-h|--help Show this message.
Examples:
mlr reorder -f a,b sends input record "d=4,b=2,a=1,c=3" to "a=1,b=2,d=4,c=3".
mlr reorder -e -f a,b sends input record "d=4,b=2,a=1,c=3" to "d=4,c=3,a=1,b=2".
This pivots specified field names to the start or end of the record -- for example when you have highly multi-column data and you want to bring a field or two to the front of line where you can give a quick visual scan.
mlr --opprint cat data/small
a b i x y pan pan 1 0.346791 0.726802 eks pan 2 0.758679 0.522151 wye wye 3 0.204603 0.338318 eks wye 4 0.381399 0.134188 wye pan 5 0.573288 0.863624
mlr --opprint reorder -f i,b data/small
i b a x y 1 pan pan 0.346791 0.726802 2 pan eks 0.758679 0.522151 3 wye wye 0.204603 0.338318 4 wye eks 0.381399 0.134188 5 pan wye 0.573288 0.863624
mlr --opprint reorder -e -f i,b data/small
a x y i b pan 0.346791 0.726802 1 pan eks 0.758679 0.522151 2 pan wye 0.204603 0.338318 3 wye eks 0.381399 0.134188 4 wye wye 0.573288 0.863624 5 pan
repeat¶
mlr repeat --help
Usage: mlr repeat [options]
Copies input records to output records multiple times.
Options must be exactly one of the following:
-n {repeat count} Repeat each input record this many times.
-f {field name} Same, but take the repeat count from the specified
field name of each input record.
-h|--help Show this message.
Example:
echo x=0 | mlr repeat -n 4 then put '$x=urand()'
produces:
x=0.488189
x=0.484973
x=0.704983
x=0.147311
Example:
echo a=1,b=2,c=3 | mlr repeat -f b
produces:
a=1,b=2,c=3
a=1,b=2,c=3
Example:
echo a=1,b=2,c=3 | mlr repeat -f c
produces:
a=1,b=2,c=3
a=1,b=2,c=3
a=1,b=2,c=3
This is useful in at least two ways: one, as a data-generator as in the
above example using urand(); two, for reconstructing individual
samples from data which has been count-aggregated:
cat data/repeat-example.dat
color=blue,count=5 color=red,count=4 color=green,count=3
mlr repeat -f count then cut -x -f count data/repeat-example.dat
color=blue color=blue color=blue color=blue color=blue color=red color=red color=red color=red color=green color=green color=green
After expansion with repeat, such data can then be sent on to
stats1 -a mode, or (if the data are numeric) to stats1 -a
p10,p50,p90, etc.
reshape¶
mlr reshape --help
Usage: mlr reshape [options]
Wide-to-long options:
-i {input field names} -o {key-field name,value-field name}
-r {input field regex} -o {key-field name,value-field name}
These pivot/reshape the input data such that the input fields are removed
and separate records are emitted for each key/value pair.
Note: if you have multiple regexes, please specify them using multiple -r,
since regexes can contain commas within them.
Note: this works with tail -f and produces output records for each input
record seen. If input is coming from `tail -f`, be sure to use
`--records-per-batch 1`.
Long-to-wide options:
-s {key-field name,value-field name}
These pivot/reshape the input data to undo the wide-to-long operation.
Note: this does not work with tail -f; it produces output records only after
all input records have been read.
Examples:
Input file "wide.txt":
time X Y
2009-01-01 0.65473572 2.4520609
2009-01-02 -0.89248112 0.2154713
2009-01-03 0.98012375 1.3179287
mlr --pprint reshape -i X,Y -o item,value wide.txt
time item value
2009-01-01 X 0.65473572
2009-01-01 Y 2.4520609
2009-01-02 X -0.89248112
2009-01-02 Y 0.2154713
2009-01-03 X 0.98012375
2009-01-03 Y 1.3179287
mlr --pprint reshape -r '[A-Z]' -o item,value wide.txt
time item value
2009-01-01 X 0.65473572
2009-01-01 Y 2.4520609
2009-01-02 X -0.89248112
2009-01-02 Y 0.2154713
2009-01-03 X 0.98012375
2009-01-03 Y 1.3179287
Input file "long.txt":
time item value
2009-01-01 X 0.65473572
2009-01-01 Y 2.4520609
2009-01-02 X -0.89248112
2009-01-02 Y 0.2154713
2009-01-03 X 0.98012375
2009-01-03 Y 1.3179287
mlr --pprint reshape -s item,value long.txt
time X Y
2009-01-01 0.65473572 2.4520609
2009-01-02 -0.89248112 0.2154713
2009-01-03 0.98012375 1.3179287
See also mlr nest.
sample¶
mlr sample --help
Usage: mlr sample [options]
Reservoir sampling (subsampling without replacement), optionally by category.
See also mlr bootstrap and mlr shuffle.
Options:
-g {a,b,c} Optional: group-by-field names for samples, e.g. a,b,c.
-k {k} Required: number of records to output in total, or by group if using -g.
-h|--help Show this message.
This is reservoir-sampling: select k items from n with
uniform probability and no repeats in the sample. (If n is less than
k, then of course only n samples are produced.) With -g
{field names}, produce a k-sample for each distinct value of the
specified field names.
$ mlr --opprint sample -k 4 data/colored-shapes.dkvp color shape flag i u v w x purple triangle 0 90122 0.9986871176198068 0.3037738877233719 0.5154934457238382 5.365962021016529 red circle 0 3139 0.04835898233323954 -0.03964684310055758 0.5263660881848111 5.3758779366493625 orange triangle 0 67847 0.36746306902109926 0.5161574810505635 0.5176199566173642 3.1748088656576567 yellow square 1 33576 0.3098376725521097 0.8525628505287842 0.49774122460981685 4.494754378604669 $ mlr --opprint sample -k 4 data/colored-shapes.dkvp color shape flag i u v w x blue square 1 16783 0.09974385090654347 0.7243899920872646 0.5353718443278438 4.431057737383438 orange square 1 93291 0.5944176543007182 0.17744449786454086 0.49262281749172077 3.1548117990710653 yellow square 1 54436 0.5268161165014636 0.8785588662666121 0.5058773791931063 7.019185838783636 yellow square 1 55491 0.0025440267883102274 0.05474106287787284 0.5102729153751984 3.526301273728043 $ mlr --opprint sample -k 2 -g color data/colored-shapes.dkvp color shape flag i u v w x yellow triangle 1 11 0.6321695890307647 0.9887207810889004 0.4364983936735774 5.7981881667050565 yellow square 1 917 0.8547010348386344 0.7356782810796262 0.4531511689924275 5.774541777078352 red circle 1 4000 0.05490416175132373 0.07392337815122155 0.49416101516594396 5.355725080701707 red square 0 87506 0.6357719216821314 0.6970867759393995 0.4940826462055272 6.351579417310387 purple triangle 0 14898 0.7800986870203719 0.23998073813992293 0.5014775988383656 3.141006771777843 purple triangle 0 151 0.032614487569017414 0.7346633365041219 0.7812143304483805 2.6831992610568047 green triangle 1 126 0.1513010528347546 0.40346767294704544 0.051213231883952326 5.955109300797182 green circle 0 17635 0.029856606049114442 0.4724542934246524 0.49529606749929744 5.239153910272168 blue circle 1 1020 0.414263129226617 0.8304946402876182 0.13151094520189244 4.397873687920433 blue triangle 0 220 0.441773289968473 0.44597731903759075 0.6329360666849821 4.3064608776550894 orange square 0 1885 0.8079311983747106 0.8685956833908394 0.3116410800256374 4.390864584500387 orange triangle 0 1533 0.32904497195507487 0.23168161807490417 0.8722623057355134 5.164071635714438 $ mlr --opprint sample -k 2 -g color then sort -f color data/colored-shapes.dkvp color shape flag i u v w x blue circle 0 215 0.7803586969333292 0.33146680638888126 0.04289047852629113 5.725365736377487 blue circle 1 3616 0.8548431579124808 0.4989623130006362 0.3339426415875795 3.696785877560498 green square 0 356 0.7674272008085286 0.341578843118008 0.4570224877870851 4.830320062215299 green square 0 152 0.6684429446914862 0.016056003736548696 0.4656148241291592 5.434588759225423 orange triangle 0 587 0.5175826237797857 0.08989091493635304 0.9011709461770973 4.265854207755811 orange triangle 0 1533 0.32904497195507487 0.23168161807490417 0.8722623057355134 5.164071635714438 purple triangle 0 14192 0.5196327866973567 0.7860928603468063 0.4964368415453642 4.899167143824484 purple triangle 0 65 0.6842806710360729 0.5823723856331258 0.8014053396013747 5.805148213865135 red square 1 2431 0.38378504852300466 0.11445015005595527 0.49355539228753786 5.146756570128739 red triangle 0 57097 0.43763430414406546 0.3355450325004481 0.5322349637512487 4.144267240289442 yellow triangle 1 11 0.6321695890307647 0.9887207810889004 0.4364983936735774 5.7981881667050565 yellow square 1 158 0.41527900739142165 0.7118027080775757 0.4200799665161291 5.33279067554884
Note that no output is produced until all inputs are in. Another way to do
sampling, which works in the streaming case, is mlr filter 'urand() &
0.001' where you tune the 0.001 to meet your needs.
sec2gmt¶
mlr sec2gmt -h
Usage: mlr sec2gmt [options] {comma-separated list of field names}
Replaces a numeric field representing seconds since the epoch with the
corresponding GMT timestamp; leaves non-numbers as-is. This is nothing
more than a keystroke-saver for the sec2gmt function:
mlr sec2gmt time1,time2
is the same as
mlr put '$time1 = sec2gmt($time1); $time2 = sec2gmt($time2)'
Options:
-1 through -9: format the seconds using 1..9 decimal places, respectively.
--millis Input numbers are treated as milliseconds since the epoch.
--micros Input numbers are treated as microseconds since the epoch.
--nanos Input numbers are treated as nanoseconds since the epoch.
-h|--help Show this message.
sec2gmtdate¶
mlr sec2gmtdate -h
Usage: ../c/mlr sec2gmtdate {comma-separated list of field names}
Replaces a numeric field representing seconds since the epoch with the
corresponding GMT year-month-day timestamp; leaves non-numbers as-is.
This is nothing more than a keystroke-saver for the sec2gmtdate function:
../c/mlr sec2gmtdate time1,time2
is the same as
../c/mlr put '$time1=sec2gmtdate($time1);$time2=sec2gmtdate($time2)'
seqgen¶
mlr seqgen -h
Usage: mlr seqgen [options]
Passes input records directly to output. Most useful for format conversion.
Produces a sequence of counters. Discards the input record stream. Produces
output as specified by the options
Options:
-f {name} (default "i") Field name for counters.
--start {value} (default 1) Inclusive start value.
--step {value} (default 1) Step value.
--stop {value} (default 100) Inclusive stop value.
-h|--help Show this message.
Start, stop, and/or step may be floating-point. Output is integer if start,
stop, and step are all integers. Step may be negative. It may not be zero
unless start == stop.
mlr seqgen --stop 10
i=1 i=2 i=3 i=4 i=5 i=6 i=7 i=8 i=9 i=10
mlr seqgen --start 20 --stop 40 --step 4
i=20 i=24 i=28 i=32 i=36 i=40
mlr seqgen --start 40 --stop 20 --step -4
i=40 i=36 i=32 i=28 i=24 i=20
shuffle¶
mlr shuffle -h
Usage: mlr shuffle [options] Outputs records randomly permuted. No output records are produced until all input records are read. See also mlr bootstrap and mlr sample. Options: -h|--help Show this message.
skip-trivial-records¶
mlr skip-trivial-records -h
Usage: mlr skip-trivial-records [options] Passes through all records except those with zero fields, or those for which all fields have empty value. Options: -h|--help Show this message.
cat data/trivial-records.csv
a,b,c 1,2,3 4,,6 ,, ,8,9
mlr --csv skip-trivial-records data/trivial-records.csv
a,b,c 1,2,3 4,,6 ,8,9
sort¶
mlr sort --help
Usage: mlr sort {flags}
Sorts records primarily by the first specified field, secondarily by the second
field, and so on. (Any records not having all specified sort keys will appear
at the end of the output, in the order they were encountered, regardless of the
specified sort order.) The sort is stable: records that compare equal will sort
in the order they were encountered in the input record stream.
Options:
-f {comma-separated field names} Lexical ascending
-r {comma-separated field names} Lexical descending
-c {comma-separated field names} Case-folded lexical ascending
-cr {comma-separated field names} Case-folded lexical descending
-n {comma-separated field names} Numerical ascending; nulls sort last
-nf {comma-separated field names} Same as -n
-nr {comma-separated field names} Numerical descending; nulls sort first
-t {comma-separated field names} Natural ascending
-tr|-rt {comma-separated field names} Natural descending
-h|--help Show this message.
Example:
mlr sort -f a,b -nr x,y,z
which is the same as:
mlr sort -f a -f b -nr x -nr y -nr z
Example:
mlr --opprint sort -f a -nr x data/small
a b i x y eks pan 2 0.758679 0.522151 eks wye 4 0.381399 0.134188 pan pan 1 0.346791 0.726802 wye pan 5 0.573288 0.863624 wye wye 3 0.204603 0.338318
Here's an example filtering log data: suppose multiple threads (labeled here by color) are all logging progress counts to a single log file. The log file is (by nature) chronological, so the progress of various threads is interleaved:
head -n 10 data/multicountdown.dat
upsec=0.002,color=green,count=1203 upsec=0.083,color=red,count=3817 upsec=0.188,color=red,count=3801 upsec=0.395,color=blue,count=2697 upsec=0.526,color=purple,count=953 upsec=0.671,color=blue,count=2684 upsec=0.899,color=purple,count=926 upsec=0.912,color=red,count=3798 upsec=1.093,color=blue,count=2662 upsec=1.327,color=purple,count=917
We can group these by thread by sorting on the thread ID (here,
color). Since Miller's sort is stable, this means that
timestamps within each thread's log data are still chronological:
head -n 20 data/multicountdown.dat | mlr --opprint sort -f color
upsec color count 0.395 blue 2697 0.671 blue 2684 1.093 blue 2662 2.064 blue 2659 2.2880000000000003 blue 2647 0.002 green 1203 1.407 green 1187 1.448 green 1177 2.313 green 1161 0.526 purple 953 0.899 purple 926 1.327 purple 917 1.703 purple 908 0.083 red 3817 0.188 red 3801 0.912 red 3798 1.416 red 3788 1.587 red 3782 1.601 red 3755 1.832 red 3717
Any records not having all specified sort keys will appear at the end of the output, in the order they were encountered, regardless of the specified sort order:
mlr sort -n x data/sort-missing.dkvp
x=1 x=2 x=4 a=3
mlr sort -nr x data/sort-missing.dkvp
x=4 x=2 x=1 a=3
sort-within-records¶
mlr sort-within-records -h
Usage: mlr sort-within-records [options] Outputs records sorted lexically ascending by keys. Options: -r Recursively sort subobjects/submaps, e.g. for JSON input. -h|--help Show this message.
cat data/sort-within-records.json
{
"a": 1,
"b": 2,
"c": 3
}
{
"b": 4,
"a": 5,
"c": 6
}
{
"c": 7,
"b": 8,
"a": 9
}
mlr --ijson --opprint cat data/sort-within-records.json
a b c 1 2 3 b a c 4 5 6 c b a 7 8 9
mlr --json sort-within-records data/sort-within-records.json
[
{
"a": 1,
"b": 2,
"c": 3
},
{
"a": 5,
"b": 4,
"c": 6
},
{
"a": 9,
"b": 8,
"c": 7
}
]
mlr --ijson --opprint sort-within-records data/sort-within-records.json
a b c 1 2 3 5 4 6 9 8 7
sparsify¶
mlr sparsify --help
Usage: mlr sparsify [options]
Unsets fields for which the key is the empty string (or, optionally, another
specified value). Only makes sense with output format not being CSV or TSV.
Options:
-s {filler string} What values to remove. Defaults to the empty string.
-f {a,b,c} Specify field names to be operated on; any other fields won't be
modified. The default is to modify all fields.
-h|--help Show this message.
Example: if input is a=1,b=,c=3 then output is a=1,c=3.
split¶
mlr split --help
Usage: mlr split [options] {filename}
Options:
-n {n}: Cap file sizes at N records.
-m {m}: Produce M files, round-robining records among them.
-g {a,b,c}: Write separate files with records having distinct values for fields named a,b,c.
Exactly one of -m, -n, or -g must be supplied.
--prefix {p} Specify filename prefix; default "split".
--suffix {s} Specify filename suffix; default is from mlr output format, e.g. "csv".
-a Append to existing file(s), if any, rather than overwriting.
-v Send records along to downstream verbs as well as splitting to files.
-e Do NOT URL-escape names of output files.
-j {J} Use string J to join filename parts; default "_".
-h|--help Show this message.
Any of the output-format command-line flags (see mlr -h). For example, using
mlr --icsv --from myfile.csv split --ojson -n 1000
the input is CSV, but the output files are JSON.
Examples: Suppose myfile.csv has 1,000,000 records.
100 output files, 10,000 records each. First 10,000 records in split_1.csv, next in split_2.csv, etc.
mlr --csv --from myfile.csv split -n 10000
10 output files, 100,000 records each. Records 1,11,21,etc in split_1.csv, records 2,12,22, etc in split_2.csv, etc.
mlr --csv --from myfile.csv split -m 10
Same, but with JSON output.
mlr --csv --from myfile.csv split -m 10 -o json
Same but instead of split_1.csv, split_2.csv, etc. there are test_1.dat, test_2.dat, etc.
mlr --csv --from myfile.csv split -m 10 --prefix test --suffix dat
Same, but written to the /tmp/ directory.
mlr --csv --from myfile.csv split -m 10 --prefix /tmp/test --suffix dat
If the shape field has values triangle and square, then there will be split_triangle.csv and split_square.csv.
mlr --csv --from myfile.csv split -g shape
If the color field has values yellow and green, and the shape field has values triangle and square,
then there will be split_yellow_triangle.csv, split_yellow_square.csv, etc.
mlr --csv --from myfile.csv split -g color,shape
See also the "tee" DSL function which lets you do more ad-hoc customization.
ssub¶
mlr ssub -h
Usage: mlr ssub [options]
Replaces old string with new string in specified field(s), without regex support for
the old string, like the `ssub` DSL function. See also the `gsub` and `sub` verbs.
Options:
-f {a,b,c} Field names to convert.
-h|--help Show this message.
mlr --icsv --opprint --from example.csv cat --filename then sub -f filename . o
filename color shape flag k index quantity rate oxample.csv yellow triangle true 1 11 43.6498 9.8870 oxample.csv red square true 2 15 79.2778 0.0130 oxample.csv red circle true 3 16 13.8103 2.9010 oxample.csv red square false 4 48 77.5542 7.4670 oxample.csv purple triangle false 5 51 81.2290 8.5910 oxample.csv red square false 6 64 77.1991 9.5310 oxample.csv purple triangle false 7 65 80.1405 5.8240 oxample.csv yellow circle true 8 73 63.9785 4.2370 oxample.csv yellow circle true 9 87 63.5058 8.3350 oxample.csv purple square false 10 91 72.3735 8.2430
mlr --icsv --opprint --from example.csv cat --filename then ssub -f filename . o
filename color shape flag k index quantity rate exampleocsv yellow triangle true 1 11 43.6498 9.8870 exampleocsv red square true 2 15 79.2778 0.0130 exampleocsv red circle true 3 16 13.8103 2.9010 exampleocsv red square false 4 48 77.5542 7.4670 exampleocsv purple triangle false 5 51 81.2290 8.5910 exampleocsv red square false 6 64 77.1991 9.5310 exampleocsv purple triangle false 7 65 80.1405 5.8240 exampleocsv yellow circle true 8 73 63.9785 4.2370 exampleocsv yellow circle true 9 87 63.5058 8.3350 exampleocsv purple square false 10 91 72.3735 8.2430
stats1¶
mlr stats1 --help
Usage: mlr stats1 [options]
Computes univariate statistics for one or more given fields, accumulated across
the input record stream.
Options:
-a {sum,count,...} Names of accumulators: one or more of:
median This is the same as p50
p10 p25.2 p50 p98 p100 etc.
count Count instances of fields
null_count Count number of empty-string/JSON-null instances per field
distinct_count Count number of distinct values per field
mode Find most-frequently-occurring values for fields; first-found wins tie
antimode Find least-frequently-occurring values for fields; first-found wins tie
sum Compute sums of specified fields
mean Compute averages (sample means) of specified fields
var Compute sample variance of specified fields
stddev Compute sample standard deviation of specified fields
meaneb Estimate error bars for averages (assuming no sample autocorrelation)
skewness Compute sample skewness of specified fields
kurtosis Compute sample kurtosis of specified fields
min Compute minimum values of specified fields
max Compute maximum values of specified fields
minlen Compute minimum string-lengths of specified fields
maxlen Compute maximum string-lengths of specified fields
-f {a,b,c} Value-field names on which to compute statistics
--fr {regex} Regex for value-field names on which to compute statistics
(compute statistics on values in all field names matching regex
--fx {regex} Inverted regex for value-field names on which to compute statistics
(compute statistics on values in all field names not matching regex)
-g {d,e,f} Optional group-by-field names
--gr {regex} Regex for optional group-by-field names
(group by values in field names matching regex)
--gx {regex} Inverted regex for optional group-by-field names
(group by values in field names not matching regex)
--grfx {regex} Shorthand for --gr {regex} --fx {that same regex}
-i Use interpolated percentiles, like R's type=7; default like type=1.
Not sensical for string-valued fields.\n");
-s Print iterative stats. Useful in tail -f contexts, in which
case please avoid pprint-format output since end of input
stream will never be seen. Likewise, if input is coming from `tail -f`
be sure to use `--records-per-batch 1`.
-h|--help Show this message.
Example: mlr stats1 -a min,p10,p50,p90,max -f value -g size,shape
Example: mlr stats1 -a count,mode -f size
Example: mlr stats1 -a count,mode -f size -g shape
Example: mlr stats1 -a count,mode --fr '^[a-h].*$' -gr '^k.*$'
This computes count and mode statistics on all field names beginning
with a through h, grouped by all field names starting with k.
Notes:
* p50 and median are synonymous.
* min and max output the same results as p0 and p100, respectively, but use
less memory.
* String-valued data make sense unless arithmetic on them is required,
e.g. for sum, mean, interpolated percentiles, etc. In case of mixed data,
numbers are less than strings.
* count and mode allow text input; the rest require numeric input.
In particular, 1 and 1.0 are distinct text for count and mode.
* When there are mode ties, the first-encountered datum wins.
These are simple univariate statistics on one or more number-valued fields
(count and mode apply to non-numeric fields as well),
optionally categorized by one or more other fields.
mlr --oxtab stats1 -a count,sum,min,p10,p50,mean,p90,max -f x,y data/medium
x_count 10000 x_sum 4986.019681679581 x_min 0.00004509679127584487 x_p10 0.09332217805283527 x_p50 0.5011592202840128 x_mean 0.49860196816795804 x_p90 0.900794437962015 x_max 0.999952670371898 y_count 10000 y_sum 5062.057444929905 y_min 0.00008818962627266114 y_p10 0.10213207378968225 y_p50 0.5060212582772865 y_mean 0.5062057444929905 y_p90 0.9053657573378745 y_max 0.9999648102177897
mlr --opprint stats1 -a mean -f x,y -g b then sort -f b data/medium
b x_mean y_mean eks 0.5063609846272304 0.510292657158104 hat 0.4878988625336502 0.5131176341556505 pan 0.4973036405471583 0.49959885012092725 wye 0.4975928392133964 0.5045964890907357 zee 0.5042419022900586 0.5029967546798116
mlr --c2p stats1 -a p50,p99 -f u,v -g color \ then put '$ur=$u_p99/$u_p50;$vr=$v_p99/$v_p50' \ data/colored-shapes.csv
color u_p50 u_p99 v_p50 v_p99 ur vr yellow 0.501019 0.989046 0.520630 0.987034 1.974068847688411 1.895845418051207 red 0.485038 0.990054 0.492586 0.994444 2.0411885254351203 2.0188231090611586 purple 0.501319 0.988893 0.504571 0.988287 1.9725823278192132 1.9586678584381585 green 0.502015 0.990764 0.505359 0.990175 1.9735744947860123 1.9593496900223406 blue 0.525226 0.992655 0.485170 0.993873 1.8899578467174132 2.048504647855391 orange 0.483548 0.993635 0.480913 0.989102 2.054883899840347 2.056717119312641
mlr --c2p count-distinct -f shape then sort -nr count data/colored-shapes.csv
shape count square 4115 triangle 3372 circle 2591
mlr --c2p stats1 -a mode -f color -g shape data/colored-shapes.csv
shape color_mode triangle red square red circle red
stats2¶
mlr stats2 --help
Usage: mlr stats2 [options]
Computes bivariate statistics for one or more given field-name pairs,
accumulated across the input record stream.
-a {linreg-ols,corr,...} Names of accumulators: one or more of:
linreg-ols Linear regression using ordinary least squares
linreg-pca Linear regression using principal component analysis
r2 Quality metric for linreg-ols (linreg-pca emits its own)
logireg Logistic regression
corr Sample correlation
cov Sample covariance
covx Sample-covariance matrix
-f {a,b,c,d} Value-field name-pairs on which to compute statistics.
There must be an even number of names.
-g {e,f,g} Optional group-by-field names.
-v Print additional output for linreg-pca.
-s Print iterative stats. Useful in tail -f contexts, in which
case please avoid pprint-format output since end of input
stream will never be seen. Likewise, if input is coming from
`tail -f`, be sure to use `--records-per-batch 1`.
--fit Rather than printing regression parameters, applies them to
the input data to compute new fit fields. All input records are
held in memory until end of input stream. Has effect only for
linreg-ols, linreg-pca, and logireg.
Only one of -s or --fit may be used.
Example: mlr stats2 -a linreg-pca -f x,y
Example: mlr stats2 -a linreg-ols,r2 -f x,y -g size,shape
Example: mlr stats2 -a corr -f x,y
These are simple bivariate statistics on one or more pairs of number-valued fields, optionally categorized by one or more fields.
mlr --oxtab put '$x2=$x*$x; $xy=$x*$y; $y2=$y**2' \ then stats2 -a cov,corr -f x,y,y,y,x2,xy,x2,y2 \ data/medium
x_y_cov 0.00004257482082749404 x_y_corr 0.0005042001844473328 y_y_cov 0.08461122467974005 y_y_corr 1 x2_xy_cov 0.041883822817793716 x2_xy_corr 0.6301743420379936 x2_y2_cov -0.0003095372596253918 x2_y2_corr -0.003424908876111875
mlr --opprint put '$x2=$x*$x; $xy=$x*$y; $y2=$y**2' \ then stats2 -a linreg-ols,r2 -f x,y,y,y,xy,y2 -g a \ data/medium
a x_y_ols_m x_y_ols_b x_y_ols_n x_y_r2 y_y_ols_m y_y_ols_b y_y_ols_n y_y_r2 xy_y2_ols_m xy_y2_ols_b xy_y2_ols_n xy_y2_r2 pan 0.017025512736819345 0.500402892289764 2081 0.00028691820445815624 1 -0.00000000000000002890430283104539 2081 1 0.8781320866715664 0.11908230147563569 2081 0.4174982737731127 eks 0.04078049236855813 0.4814020796765104 1965 0.0016461239223448218 1 0.00000000000000017862676354313703 1965 1 0.897872861169018 0.1073405443361234 1965 0.4556322386425451 wye -0.03915349075204785 0.5255096523974457 1966 0.0015051268704373377 1 0.00000000000000004464425401127647 1966 1 0.8538317334220837 0.1267454301662969 1966 0.3899172181859931 zee 0.0027812364960401333 0.5043070448033061 2047 0.000007751652858787357 1 0.00000000000000004819404567023685 2047 1 0.8524439912011011 0.12401684308018947 2047 0.39356598090006495 hat -0.018620577041095272 0.5179005397264937 1941 0.00035200366460556604 1 -0.00000000000000003400445761787692 1941 1 0.8412305086345017 0.13557328318623207 1941 0.3687944261732266
Here's an example simple line-fit. The x and y
fields of the data/medium dataset are just independent uniformly
distributed on the unit interval. Here we remove half the data and fit a line to it.
# Prepare input data: mlr filter '($x<.5 && $y<.5) || ($x>.5 && $y>.5)' data/medium > data/medium-squares # Do a linear regression and examine coefficients: mlr --ofs newline stats2 -a linreg-pca -f x,y data/medium-squares x_y_pca_m=1.014419 x_y_pca_b=0.000308 x_y_pca_quality=0.861354 # Option 1 to apply the regression coefficients and produce a linear fit: # Set x_y_pca_m and x_y_pca_b as shell variables: eval $(mlr --ofs newline stats2 -a linreg-pca -f x,y data/medium-squares) # In addition to x and y, make a new yfit which is the line fit, then plot # using your favorite tool: mlr --onidx put '$yfit='$x_y_pca_m'*$x+'$x_y_pca_b then cut -x -f a,b,i data/medium-squares \ | pgr -p -title 'linreg-pca example' -xmin 0 -xmax 1 -ymin 0 -ymax 1 # Option 2 to apply the regression coefficients and produce a linear fit: use --fit option mlr --onidx stats2 -a linreg-pca --fit -f x,y then cut -f a,b,i data/medium-squares \ | pgr -p -title 'linreg-pca example' -xmin 0 -xmax 1 -ymin 0 -ymax 1
I use pgr for plotting; here's a screenshot.
(Thanks Drew Kunas for a good conversation about PCA!)
Here's an example estimating time-to-completion for a set of jobs. Input data comes from a log file, with number of work units left to do in the count field and accumulated seconds in the upsec field, labeled by the color field:
head -n 10 data/multicountdown.dat
upsec=0.002,color=green,count=1203 upsec=0.083,color=red,count=3817 upsec=0.188,color=red,count=3801 upsec=0.395,color=blue,count=2697 upsec=0.526,color=purple,count=953 upsec=0.671,color=blue,count=2684 upsec=0.899,color=purple,count=926 upsec=0.912,color=red,count=3798 upsec=1.093,color=blue,count=2662 upsec=1.327,color=purple,count=917
We can do a linear regression on count remaining as a function of time: with c = m*u+b we want to find the time when the count goes to zero, i.e. u=-b/m.
mlr --oxtab stats2 -a linreg-pca -f upsec,count -g color \ then put '$donesec = -$upsec_count_pca_b/$upsec_count_pca_m' \ data/multicountdown.dat
color green upsec_count_pca_m -32.75691673397728 upsec_count_pca_b 1213.7227296044375 upsec_count_pca_n 24 upsec_count_pca_quality 0.9999839351341062 donesec 37.052410624028525 color red upsec_count_pca_m -37.367646434187435 upsec_count_pca_b 3810.1334002923936 upsec_count_pca_n 30 upsec_count_pca_quality 0.9999894618183773 donesec 101.9634299688333 color blue upsec_count_pca_m -29.2312120633493 upsec_count_pca_b 2698.9328203182517 upsec_count_pca_n 25 upsec_count_pca_quality 0.9999590846136102 donesec 92.33051350964094 color purple upsec_count_pca_m -39.030097447953594 upsec_count_pca_b 979.9883413064917 upsec_count_pca_n 21 upsec_count_pca_quality 0.9999908956206317 donesec 25.108529196302943
step¶
mlr step --help
Usage: mlr step [options]
Computes values dependent on earlier/later records, optionally grouped by category.
Options:
-a {delta,rsum,...} Names of steppers: comma-separated, one or more of:
counter Count instances of field(s) between successive records
delta Compute differences in field(s) between successive records
ewma Exponentially weighted moving average over successive records
from-first Compute differences in field(s) from first record
ratio Compute ratios in field(s) between successive records
rprod Compute running products of field(s) between successive records
rsum Compute running sums of field(s) between successive records
shift Alias for shift_lag
shift_lag Include value(s) in field(s) from the previous record, if any
shift_lead Include value(s) in field(s) from the next record, if any
slwin Sliding-window averages over m records back and n forward. E.g. slwin_7_2 for 7 back and 2 forward.
-f {a,b,c} Value-field names on which to compute statistics
-g {d,e,f} Optional group-by-field names
-F Computes integerable things (e.g. counter) in floating point.
As of Miller 6 this happens automatically, but the flag is accepted
as a no-op for backward compatibility with Miller 5 and below.
-d {x,y,z} Weights for EWMA. 1 means current sample gets all weight (no
smoothing), near under 1 is light smoothing, near over 0 is
heavy smoothing. Multiple weights may be specified, e.g.
"mlr step -a ewma -f sys_load -d 0.01,0.1,0.9". Default if omitted
is "-d 0.5".
-o {a,b,c} Custom suffixes for EWMA output fields. If omitted, these default to
the -d values. If supplied, the number of -o values must be the same
as the number of -d values.
-h|--help Show this message.
Examples:
mlr step -a rsum -f request_size
mlr step -a delta -f request_size -g hostname
mlr step -a ewma -d 0.1,0.9 -f x,y
mlr step -a ewma -d 0.1,0.9 -o smooth,rough -f x,y
mlr step -a ewma -d 0.1,0.9 -o smooth,rough -f x,y -g group_name
mlr step -a slwin_9_0,slwin_0_9 -f x
Please see https://miller.readthedocs.io/en/latest/reference-verbs.html#filter or
https://en.wikipedia.org/wiki/Moving_average#Exponential_moving_average
for more information on EWMA.
Most Miller commands are record-at-a-time, with the exception of stats1, stats2, and histogram which compute aggregate output. The step command is intermediate: it allows the option of adding fields which are functions of fields from previous records. Rsum is short for running sum.
mlr --opprint step -a shift,delta,rsum,counter -f x data/medium | head -15
a b i x y x_shift x_delta x_rsum x_counter pan pan 1 0.3467901443380824 0.7268028627434533 - 0 0.3467901443380824 1 eks pan 2 0.7586799647899636 0.5221511083334797 0.3467901443380824 0.41188982045188116 1.105470109128046 2 wye wye 3 0.20460330576630303 0.33831852551664776 0.7586799647899636 -0.5540766590236605 1.3100734148943491 3 eks wye 4 0.38139939387114097 0.13418874328430463 0.20460330576630303 0.17679608810483793 1.6914728087654902 4 wye pan 5 0.5732889198020006 0.8636244699032729 0.38139939387114097 0.19188952593085962 2.264761728567491 5 zee pan 6 0.5271261600918548 0.49322128674835697 0.5732889198020006 -0.04616275971014583 2.7918878886593457 6 eks zee 7 0.6117840605678454 0.1878849191181694 0.5271261600918548 0.08465790047599064 3.403671949227191 7 zee wye 8 0.5985540091064224 0.976181385699006 0.6117840605678454 -0.013230051461422976 4.0022259583336135 8 hat wye 9 0.03144187646093577 0.7495507603507059 0.5985540091064224 -0.5671121326454867 4.033667834794549 9 pan wye 10 0.5026260055412137 0.9526183602969864 0.03144187646093577 0.47118412908027796 4.536293840335763 10 pan pan 11 0.7930488423451967 0.6505816637259333 0.5026260055412137 0.29042283680398295 5.32934268268096 11 zee pan 12 0.3676141320555616 0.23614420670296965 0.7930488423451967 -0.4254347102896351 5.696956814736522 12 eks pan 13 0.4915175580479536 0.7709126592971468 0.3676141320555616 0.12390342599239201 6.1884743727844755 13 eks zee 14 0.5207382318405251 0.34141681118811673 0.4915175580479536 0.02922067379257154 6.709212604625001 14
mlr --opprint step -a shift,delta,rsum,counter -f x -g a data/medium | head -15
a b i x y x_shift x_delta x_rsum x_counter pan pan 1 0.3467901443380824 0.7268028627434533 - 0 0.3467901443380824 1 eks pan 2 0.7586799647899636 0.5221511083334797 - 0 0.7586799647899636 1 wye wye 3 0.20460330576630303 0.33831852551664776 - 0 0.20460330576630303 1 eks wye 4 0.38139939387114097 0.13418874328430463 0.7586799647899636 -0.3772805709188226 1.1400793586611044 2 wye pan 5 0.5732889198020006 0.8636244699032729 0.20460330576630303 0.36868561403569755 0.7778922255683036 2 zee pan 6 0.5271261600918548 0.49322128674835697 - 0 0.5271261600918548 1 eks zee 7 0.6117840605678454 0.1878849191181694 0.38139939387114097 0.23038466669670443 1.75186341922895 3 zee wye 8 0.5985540091064224 0.976181385699006 0.5271261600918548 0.07142784901456767 1.1256801691982772 2 hat wye 9 0.03144187646093577 0.7495507603507059 - 0 0.03144187646093577 1 pan wye 10 0.5026260055412137 0.9526183602969864 0.3467901443380824 0.1558358612031313 0.8494161498792961 2 pan pan 11 0.7930488423451967 0.6505816637259333 0.5026260055412137 0.29042283680398295 1.6424649922244927 3 zee pan 12 0.3676141320555616 0.23614420670296965 0.5985540091064224 -0.23093987705086083 1.4932943012538389 3 eks pan 13 0.4915175580479536 0.7709126592971468 0.6117840605678454 -0.1202665025198918 2.2433809772769036 4 eks zee 14 0.5207382318405251 0.34141681118811673 0.4915175580479536 0.02922067379257154 2.7641192091174287 5
mlr --opprint step -a ewma -f x -d 0.1,0.9 data/medium | head -15
a b i x y x_ewma_0.1 x_ewma_0.9 pan pan 1 0.3467901443380824 0.7268028627434533 0.3467901443380824 0.3467901443380824 eks pan 2 0.7586799647899636 0.5221511083334797 0.3879791263832706 0.7174909827447755 wye wye 3 0.20460330576630303 0.33831852551664776 0.36964154432157387 0.25589207346415027 eks wye 4 0.38139939387114097 0.13418874328430463 0.37081732927653055 0.3688486618304419 wye pan 5 0.5732889198020006 0.8636244699032729 0.3910644883290776 0.5528448940048447 zee pan 6 0.5271261600918548 0.49322128674835697 0.4046706555053553 0.5296980334831537 eks zee 7 0.6117840605678454 0.1878849191181694 0.4253819960116043 0.6035754578593763 zee wye 8 0.5985540091064224 0.976181385699006 0.44269919732108615 0.5990561539817179 hat wye 9 0.03144187646093577 0.7495507603507059 0.40157346523507115 0.08820330421301396 pan wye 10 0.5026260055412137 0.9526183602969864 0.41167871926568544 0.46118373540839375 pan pan 11 0.7930488423451967 0.6505816637259333 0.44981573157363663 0.7598623316515164 zee pan 12 0.3676141320555616 0.23614420670296965 0.4415955716218291 0.4068389520151571 eks pan 13 0.4915175580479536 0.7709126592971468 0.4465877702644416 0.48304969744467396 eks zee 14 0.5207382318405251 0.34141681118811673 0.4540028164220499 0.51696937840094
mlr --opprint step -a ewma -f x -d 0.1,0.9 -o smooth,rough data/medium | head -15
a b i x y x_ewma_smooth x_ewma_rough pan pan 1 0.3467901443380824 0.7268028627434533 0.3467901443380824 0.3467901443380824 eks pan 2 0.7586799647899636 0.5221511083334797 0.3879791263832706 0.7174909827447755 wye wye 3 0.20460330576630303 0.33831852551664776 0.36964154432157387 0.25589207346415027 eks wye 4 0.38139939387114097 0.13418874328430463 0.37081732927653055 0.3688486618304419 wye pan 5 0.5732889198020006 0.8636244699032729 0.3910644883290776 0.5528448940048447 zee pan 6 0.5271261600918548 0.49322128674835697 0.4046706555053553 0.5296980334831537 eks zee 7 0.6117840605678454 0.1878849191181694 0.4253819960116043 0.6035754578593763 zee wye 8 0.5985540091064224 0.976181385699006 0.44269919732108615 0.5990561539817179 hat wye 9 0.03144187646093577 0.7495507603507059 0.40157346523507115 0.08820330421301396 pan wye 10 0.5026260055412137 0.9526183602969864 0.41167871926568544 0.46118373540839375 pan pan 11 0.7930488423451967 0.6505816637259333 0.44981573157363663 0.7598623316515164 zee pan 12 0.3676141320555616 0.23614420670296965 0.4415955716218291 0.4068389520151571 eks pan 13 0.4915175580479536 0.7709126592971468 0.4465877702644416 0.48304969744467396 eks zee 14 0.5207382318405251 0.34141681118811673 0.4540028164220499 0.51696937840094
Example deriving uptime-delta from system uptime:
$ each 10 uptime | mlr -p step -a delta -f 11 ... 20:08 up 36 days, 10:38, 5 users, load averages: 1.42 1.62 1.73 0.000000 20:08 up 36 days, 10:38, 5 users, load averages: 1.55 1.64 1.74 0.020000 20:08 up 36 days, 10:38, 7 users, load averages: 1.58 1.65 1.74 0.010000 20:08 up 36 days, 10:38, 9 users, load averages: 1.78 1.69 1.76 0.040000 20:08 up 36 days, 10:39, 9 users, load averages: 2.12 1.76 1.78 0.070000 20:08 up 36 days, 10:39, 9 users, load averages: 2.51 1.85 1.81 0.090000 20:08 up 36 days, 10:39, 8 users, load averages: 2.79 1.92 1.83 0.070000 20:08 up 36 days, 10:39, 4 users, load averages: 2.64 1.90 1.83 -0.020000
sub¶
mlr sub -h
Usage: mlr sub [options]
Replaces old string with new string in specified field(s), with regex support
for the old string and not handling multiple matches, like the `sub` DSL function.
See also the `gsub` and `ssub` verbs.
Options:
-f {a,b,c} Field names to convert.
-h|--help Show this message.
mlr --icsv --opprint --from example.csv cat --filename then sub -f color,shape l X
filename color shape flag k index quantity rate example.csv yeXlow triangXe true 1 11 43.6498 9.8870 example.csv red square true 2 15 79.2778 0.0130 example.csv red circXe true 3 16 13.8103 2.9010 example.csv red square false 4 48 77.5542 7.4670 example.csv purpXe triangXe false 5 51 81.2290 8.5910 example.csv red square false 6 64 77.1991 9.5310 example.csv purpXe triangXe false 7 65 80.1405 5.8240 example.csv yeXlow circXe true 8 73 63.9785 4.2370 example.csv yeXlow circXe true 9 87 63.5058 8.3350 example.csv purpXe square false 10 91 72.3735 8.2430
mlr --icsv --opprint --from example.csv cat --filename then gsub -f color,shape l X
filename color shape flag k index quantity rate example.csv yeXXow triangXe true 1 11 43.6498 9.8870 example.csv red square true 2 15 79.2778 0.0130 example.csv red circXe true 3 16 13.8103 2.9010 example.csv red square false 4 48 77.5542 7.4670 example.csv purpXe triangXe false 5 51 81.2290 8.5910 example.csv red square false 6 64 77.1991 9.5310 example.csv purpXe triangXe false 7 65 80.1405 5.8240 example.csv yeXXow circXe true 8 73 63.9785 4.2370 example.csv yeXXow circXe true 9 87 63.5058 8.3350 example.csv purpXe square false 10 91 72.3735 8.2430
summary¶
mlr summary --help
Usage: mlr summary [options]
Show summary statistics about the input data.
All summarizers:
field_type string, int, etc. -- if a column has mixed types, all encountered types are printed
count +1 for every instance of the field across all records in the input record stream
null_count count of field values either empty string or JSON null
distinct_count count of distinct values for the field
mode most-frequently-occurring value for the field
sum sum of field values
mean mean of the field values
stddev standard deviation of the field values
var variance of the field values
skewness skewness of the field values
minlen length of shortest string representation for the field
maxlen length of longest string representation for the field
min minimum field value
p25 first-quartile field value
median median field value
p75 third-quartile field value
max maximum field value
iqr interquartile range: p75 - p25
lof lower outer fence: p25 - 3.0 * iqr
lif lower inner fence: p25 - 1.5 * iqr
uif upper inner fence: p75 + 1.5 * iqr
uof upper outer fence: p75 + 3.0 * iqr
Default summarizers:
field_type count mean min max null_count distinct_count
Notes:
* min, p25, median, p75, and max work for strings as well as numbers
* Distinct-counts are computed on string representations -- so 4.1 and 4.10 are counted as distinct here.
* If the mode is not unique in the input data, the first-encountered value is reported as the mode.
Options:
-a {mean,sum,etc.} Use only the specified summarizers.
-x {mean,sum,etc.} Use all summarizers, except the specified ones.
--all Use all available summarizers.
-h|--help Show this message.
mlr --ofmt %.3f --from data/medium --opprint summary
field_name field_type count null_count distinct_count mean min max a string 10000 0 5 - eks zee b string 10000 0 5 - eks zee i int 10000 0 10000 5000.500 1 10000 x float 10000 0 10000 0.499 0.000 1.000 y float 10000 0 10000 0.506 0.000 1.000
mlr --from data/medium --opprint summary --transpose --all
field_name a b i x y field_type string string int float float count 10000 10000 10000 10000 10000 null_count 0 0 0 0 0 distinct_count 5 5 10000 10000 10000 mode pan wye 1 0.3467901443380824 0.7268028627434533 sum 0 0 50005000 4986.019681679581 5062.057444929905 mean - - 5000.5 0.49860196816795804 0.5062057444929905 stddev - - 2886.8956799071675 0.29029251511440074 0.2908800864269331 var - - 8334166.666666667 0.08426974433144457 0.08461122467974005 skewness - - 0 -0.0006899591185517494 -0.01784976012013298 minlen 3 3 1 15 13 maxlen 3 3 5 22 22 min eks eks 1 0.00004509679127584487 0.00008818962627266114 p25 hat hat 2501 0.24667037823231752 0.25213670524015686 median pan pan 5001 0.5011592202840128 0.5060212582772865 p75 wye wye 7501 0.7481860062358446 0.7640028449996572 max zee zee 10000 0.999952670371898 0.9999648102177897 iqr - - 5000 0.5015156280035271 0.5118661397595003 lof - - -12499 -1.2578765057782637 -1.2834617140383442 lif - - -4999 -0.5056030637729731 -0.5156625043990937 uif - - 15001 1.5004594482411353 1.5318020546389077 uof - - 22501 2.252732890246426 2.2996012642781585
mlr --from data/medium --opprint summary --transpose -a mean,median,mode
field_name a b i x y mode pan wye 1 0.3467901443380824 0.7268028627434533 mean - - 5000.5 0.49860196816795804 0.5062057444929905 median pan pan 5001 0.5011592202840128 0.5060212582772865
tac¶
mlr tac --help
Usage: mlr tac [options] Prints records in reverse order from the order in which they were encountered. Options: -h|--help Show this message.
Prints the records in the input stream in reverse order. Note: this requires Miller to retain all input records in memory before any output records are produced.
mlr --icsv --opprint cat data/a.csv
a b c 1 2 3 4 5 6
mlr --icsv --opprint cat data/b.csv
a b c 7 8 9
mlr --icsv --opprint tac data/a.csv data/b.csv
a b c 7 8 9 4 5 6 1 2 3
mlr --icsv --opprint put '$filename=FILENAME' then tac data/a.csv data/b.csv
a b c filename 7 8 9 data/b.csv 4 5 6 data/a.csv 1 2 3 data/a.csv
tail¶
mlr tail --help
Usage: mlr tail [options]
Passes through the last n records, optionally by category.
Options:
-g {a,b,c} Optional group-by-field names for head counts, e.g. a,b,c.
-n {n} Head-count to print. Default 10.
-h|--help Show this message.
Prints the last n records in the input stream, optionally by category.
mlr --c2p tail -n 4 data/colored-shapes.csv
color shape flag i u v w x blue square 1 499872 0.618906 0.263796 0.531147 6.210738 blue triangle 0 499880 0.008111 0.826727 0.473296 6.146957 yellow triangle 0 499955 0.383942 0.559529 0.511376 4.307974 yellow circle 1 499974 0.764951 0.252842 0.499699 5.013810
mlr --c2p tail -n 1 -g shape data/colored-shapes.csv
color shape flag i u v w x yellow triangle 0 499955 0.383942 0.559529 0.511376 4.307974 blue square 1 499872 0.618906 0.263796 0.531147 6.210738 yellow circle 1 499974 0.764951 0.252842 0.499699 5.013810
tee¶
mlr tee --help
Usage: mlr tee [options] {filename}
Options:
-a Append to existing file, if any, rather than overwriting.
-p Treat filename as a pipe-to command.
Any of the output-format command-line flags (see mlr -h). Example: using
mlr --icsv --opprint put '...' then tee --ojson ./mytap.dat then stats1 ...
the input is CSV, the output is pretty-print tabular, but the tee-file output
is written in JSON format.
-h|--help Show this message.
template¶
mlr template --help
Usage: mlr template [options]
Places input-record fields in the order specified by list of column names.
If the input record is missing a specified field, it will be filled with the fill-with.
If the input record possesses an unspecified field, it will be discarded.
Options:
-f {a,b,c} Comma-separated field names for template, e.g. a,b,c.
-t {filename} CSV file whose header line will be used for template.
--fill-with {filler string} What to fill absent fields with. Defaults to the empty string.
-h|--help Show this message.
Example:
* Specified fields are a,b,c.
* Input record is c=3,a=1,f=6.
* Output record is a=1,b=,c=3.
top¶
mlr top --help
Usage: mlr top [options]
-f {a,b,c} Value-field names for top counts.
-g {d,e,f} Optional group-by-field names for top counts.
-n {count} How many records to print per category; default 1.
-a Print all fields for top-value records; default is
to print only value and group-by fields. Requires a single
value-field name only.
--min Print top smallest values; default is top largest values.
-F Keep top values as floats even if they look like integers.
-o {name} Field name for output indices. Default "top_idx".
This is ignored if -a is used.
Prints the n records with smallest/largest values at specified fields,
optionally by category. If -a is given, then the top records are emitted
with the same fields as they appeared in the input. Without -a, only fields
from -f, fields from -g, and the top-index field are emitted. For more information
please see https://miller.readthedocs.io/en/latest/reference-verbs#top
Note that top is distinct from head -- head shows fields which appear first in the data stream; top shows fields which are numerically largest (or smallest).
mlr --c2p cat example.csv
color shape flag k index quantity rate yellow triangle true 1 11 43.6498 9.8870 red square true 2 15 79.2778 0.0130 red circle true 3 16 13.8103 2.9010 red square false 4 48 77.5542 7.4670 purple triangle false 5 51 81.2290 8.5910 red square false 6 64 77.1991 9.5310 purple triangle false 7 65 80.1405 5.8240 yellow circle true 8 73 63.9785 4.2370 yellow circle true 9 87 63.5058 8.3350 purple square false 10 91 72.3735 8.2430
mlr --c2p top -n 1 -f quantity example.csv
top_idx quantity_top 1 81.2290
mlr --c2p top -n 1 -f quantity -g shape example.csv
shape top_idx quantity_top triangle 1 81.2290 square 1 79.2778 circle 1 63.9785
mlr --c2p top -n 1 -f quantity -g shape -o someothername example.csv
shape someothername quantity_top triangle 1 81.2290 square 1 79.2778 circle 1 63.9785
mlr --c2p top -n 1 -f quantity -g shape -a example.csv
color shape flag k index quantity rate purple triangle false 5 51 81.2290 8.5910 red square true 2 15 79.2778 0.0130 yellow circle true 8 73 63.9785 4.2370
mlr --c2p top -n 1 -f quantity -g shape -a then sort -f shape example.csv
color shape flag k index quantity rate yellow circle true 8 73 63.9785 4.2370 red square true 2 15 79.2778 0.0130 purple triangle false 5 51 81.2290 8.5910
unflatten¶
mlr unflatten --help
Usage: mlr unflatten [options]
Reverses flatten. Example: field with name 'a.b.c' and value 4
becomes name 'a' and value '{"b": { "c": 4 }}'.
Options:
-f {a,b,c} Comma-separated list of field names to unflatten (default all).
-s {string} Separator, defaulting to mlr --flatsep value.
-h|--help Show this message.
uniq¶
mlr uniq --help
Usage: mlr uniq [options]
Prints distinct values for specified field names. With -c, same as
count-distinct. For uniq, -f is a synonym for -g.
Options:
-g {d,e,f} Group-by-field names for uniq counts.
-x {a,b,c} Field names to exclude for uniq: use each record's others instead.
-c Show repeat counts in addition to unique values.
-n Show only the number of distinct values.
-o {name} Field name for output count. Default "count".
-a Output each unique record only once. Incompatible with -g.
With -c, produces unique records, with repeat counts for each.
With -n, produces only one record which is the unique-record count.
With neither -c nor -n, produces unique records.
There are two main ways to use mlr uniq: the first way is with -g to specify group-by columns.
wc -l data/colored-shapes.csv
10079 data/colored-shapes.csv
mlr --csv uniq -g color,shape data/colored-shapes.csv
color,shape yellow,triangle red,square red,circle purple,triangle yellow,circle purple,square yellow,square red,triangle green,triangle green,square blue,circle blue,triangle purple,circle blue,square green,circle orange,triangle orange,square orange,circle
mlr --c2p uniq -g color,shape -c then sort -f color,shape data/colored-shapes.csv
color shape count blue circle 384 blue square 589 blue triangle 497 green circle 287 green square 454 green triangle 368 orange circle 68 orange square 128 orange triangle 107 purple circle 289 purple square 481 purple triangle 372 red circle 1207 red square 1874 red triangle 1560 yellow circle 356 yellow square 589 yellow triangle 468
mlr --c2p uniq -g color,shape -c -o someothername \ then sort -nr someothername \ data/colored-shapes.csv
color shape someothername red square 1874 red triangle 1560 red circle 1207 yellow square 589 blue square 589 blue triangle 497 purple square 481 yellow triangle 468 green square 454 blue circle 384 purple triangle 372 green triangle 368 yellow circle 356 purple circle 289 green circle 287 orange square 128 orange triangle 107 orange circle 68
mlr --c2p uniq -n -g color,shape data/colored-shapes.csv
count 18
The second main way to use mlr uniq is without group-by columns, using -a instead:
cat data/repeats.dkvp
color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=circle,flag=1 color=red,shape=square,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=square,flag=0 color=red,shape=square,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=circle,flag=1 color=yellow,shape=circle,flag=1 color=yellow,shape=circle,flag=1 color=purple,shape=triangle,flag=0 color=yellow,shape=circle,flag=1 color=yellow,shape=circle,flag=1 color=red,shape=circle,flag=1 color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=square,flag=0 color=purple,shape=square,flag=0 color=red,shape=square,flag=0 color=red,shape=square,flag=1 color=red,shape=square,flag=0 color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=red,shape=square,flag=0 color=red,shape=square,flag=0 color=purple,shape=square,flag=0 color=red,shape=square,flag=0 color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=yellow,shape=triangle,flag=1 color=purple,shape=square,flag=0 color=yellow,shape=circle,flag=1 color=purple,shape=triangle,flag=0 color=red,shape=circle,flag=1 color=purple,shape=triangle,flag=0 color=purple,shape=triangle,flag=0 color=red,shape=square,flag=0 color=red,shape=circle,flag=1 color=red,shape=square,flag=1 color=red,shape=square,flag=0 color=red,shape=circle,flag=1 color=purple,shape=square,flag=0 color=purple,shape=square,flag=0 color=red,shape=square,flag=1 color=purple,shape=triangle,flag=0 color=purple,shape=triangle,flag=0 color=purple,shape=square,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=square,flag=0 color=yellow,shape=triangle,flag=1 color=yellow,shape=circle,flag=1 color=purple,shape=square,flag=0
wc -l data/repeats.dkvp
57 data/repeats.dkvp
mlr --opprint uniq -a data/repeats.dkvp
color shape flag red square 0 purple triangle 0 yellow circle 1 red circle 1 purple square 0 red square 1 yellow triangle 1
mlr --opprint uniq -a -n data/repeats.dkvp
count 7
mlr --opprint uniq -a -c data/repeats.dkvp
count color shape flag 17 red square 0 11 purple triangle 0 11 yellow circle 1 6 red circle 1 7 purple square 0 3 red square 1 2 yellow triangle 1
unspace¶
mlr unspace --help
Usage: mlr unspace [options]
Replaces spaces in record keys and/or values with _. This is helpful for PPRINT output.
Options:
-f {x} Replace spaces with specified filler character.
-k Unspace only keys, not keys and values.
-v Unspace only values, not keys and values.
-h|--help Show this message.
The primary use-case is for PPRINT output, which is space-delimited. For example:
cat data/spaces.csv
column 1,column 2,column 3 apple,ball,cat dale egg,fish,gale
mlr --icsv --opprint cat data/spaces.csv
column 1 column 2 column 3 apple ball cat dale egg fish gale
mlr --icsv --opprint cat data/spaces.csv
column 1 column 2 column 3 apple ball cat dale egg fish gale
mlr --icsv --opprint unspace data/spaces.csv
column_1 column_2 column_3 apple ball cat dale_egg fish gale
mlr --icsv --opprint unspace data/spaces.csv | mlr --ipprint --oxtab cat
column_1 apple column_2 ball column_3 cat column_1 dale_egg column_2 fish column_3 gale
unsparsify¶
mlr unsparsify --help
Usage: mlr unsparsify [options]
Prints records with the union of field names over all input records.
For field names absent in a given record but present in others, fills in
a value. This verb retains all input before producing any output.
Options:
--fill-with {filler string} What to fill absent fields with. Defaults to
the empty string.
-f {a,b,c} Specify field names to be operated on. Any other fields won't be
modified, and operation will be streaming.
-h|--help Show this message.
Example: if the input is two records, one being 'a=1,b=2' and the other
being 'b=3,c=4', then the output is the two records 'a=1,b=2,c=' and
'a=,b=3,c=4'.
Examples:
cat data/sparse.json
{"a":1,"b":2,"v":3}
{"u":1,"b":2}
{"a":1,"v":2,"x":3}
{"v":1,"w":2}
mlr --json unsparsify data/sparse.json
[
{
"a": 1,
"b": 2,
"v": 3,
"u": "",
"x": "",
"w": ""
},
{
"a": "",
"b": 2,
"v": "",
"u": 1,
"x": "",
"w": ""
},
{
"a": 1,
"b": "",
"v": 2,
"u": "",
"x": 3,
"w": ""
},
{
"a": "",
"b": "",
"v": 1,
"u": "",
"x": "",
"w": 2
}
]
mlr --ijson --opprint unsparsify data/sparse.json
a b v u x w 1 2 3 - - - - 2 - 1 - - 1 - 2 - 3 - - - 1 - - 2
mlr --ijson --opprint unsparsify --fill-with missing data/sparse.json
a b v u x w 1 2 3 missing missing missing missing 2 missing 1 missing missing 1 missing 2 missing 3 missing missing missing 1 missing missing 2
mlr --ijson --opprint unsparsify -f a,b,u data/sparse.json
a b v u 1 2 3 - u b a 1 2 - a v x b u 1 2 3 - - v w a b u 1 2 - - -
mlr --ijson --opprint unsparsify -f a,b,u,v,w,x then regularize data/sparse.json
a b v u w x 1 2 3 - - - - 2 - 1 - - 1 - 2 - - 3 - - 1 - 2 -